Abstract
Here, we present an update of Breast Cancer Now Tissue Bank bioinformatics, a rich platform for the sharing, mining, integration and analysis of breast cancer data. Its modalities provide researchers with access to a centralised information gateway from which they can access a network of bioinformatic resources to query findings from publicly available, in-house and experimental data generated using samples supplied from the Breast Cancer Now Tissue Bank. This in silico environment aims to help researchers use breast cancer data to their full potential, irrespective of any bioinformatics barriers. For this new release, a complete overhaul of the IT and bioinformatic infrastructure underlying the portal has been conducted and a host of novel analytical modules established. We developed and adopted an automated data selection and prioritisation system, expanded the data content and included tissue and cell line data generated from The Cancer Genome Atlas and the Cancer Cell Line Encyclopedia, designed a host of novel analytical modalities and enhanced the query building process. Furthermore, the results are presented in an interactive format, providing researchers with greater control over the information on which they want to focus. Breast Cancer Now Tissue Bank bioinformatics can be accessed at http://bioinformatics.breastcancertissuebank.org/.
INTRODUCTION
Rapid advances in technology in conjunction with their application to national and international cancer projects allows for a formidable amount of biological data to be generated at an unprecedented rate. As such, the importance of tissue banking to precision medicine has never been more relevant to cancer research than it is today. It is vital that the cancer research community has access to well-annotated tissues and the tools it requires to analyse data generated alongside the data that is accumulating in the public domain to translate these findings into personalised diagnostic, prognostic and therapeutic strategies.
The Breast Cancer Now Tissue Bank (BCNTB; http://breastcancertissuebank.org) was established in 2010 in response to a GAP analysis that highlighted a void in high-quality and fully annotated breast tissues in cancer research (1). Presented to researchers in the UK and Ireland in 2012, the BCNTB provides access to over 48,000 samples from breast tissue and blood/blood derivatives, and offers a bespoke cell line service.
Building a biobank is a notable feat but to ensure that our understanding of breast cancer remains abreast with advances in technology, we need to unite disparate fields of research and build a compendium of science from which researchers are able to address all of their needs. The aim is to create an in silico environment that has a symbiotic relationship with the in vivo and in vitro components of its tissue bank.
The BCNTB bioinformatics portal (BCNTBbp) was the first step in this process (2). This initiative was developed for the Bank—specifically as a portal from which researchers could mine and analyse breast cancer data. It provided researchers with the ability to mine literature data and access basic transcriptomic analyses conducted on publicly available -omics data. Here we describe the next evolutionary stage of this in silico resource, in which the BCNTBbp transitions from a stand-alone repository to an integrated research platform.
Since our last release, the bioinformatic infrastructure of the portal has been redesigned to accommodate the growth of data and analytical modalities offered by the Platform, and lay the foundations for links to specimen information available from the BCNTB. Other key developments include implementing novel analytical modalities, capable of analysing and integrating data generated using a range of technologies, adopting an automated system for data selection and retrieval, and enhancing the data content as well as the query-building process. Finally, we set the framework to allow researchers to analyse and integrate findings from publicly available data, in-house data and experimental data generated using samples supplied from the biobank. With the niches of this biobanking ecosystem working together seamlessly, the patient information available from the BCNTB will be updated continuously to include the molecular data generated by BCNTB bioinformatics. This will increase the dimensionality of the data available to future researchers.
RECENT DEVELOPMENTS
BCNTB:Analytics
Automated data selection, prioritisation and retrieval system
BCNTB:Analytics has adopted the Smart Automatic Classification system (SMAC; Pirrò et al., manuscript in preparation) to select, prioritise and retrieve datasets. Developed using Ruby, SMAC conducts a text search of titles and abstracts in PubMed using Boolean logical operators and retrieves documents as single XML-format files. The articles extracted are enriched by Medical Subject Headings (MeSH) terms and ranked according to a weighted score based on the total number of citations in PubMed, the abundance in breast cancer-related articles and the MeSH tree hierarchical level. A robust rank aggregation (RRA) algorithm (3), which assumes the null distribution of all the rank ratios as uniform on the unit interval, is applied to the ranked articles to ensure that statistically-relevant papers are prioritised in the analytical workflow. At the most basic level, these relationships can be viewed as cross-references between an established controlled vocabulary and the content of the paper. For each publication that fulfils the criteria, the PubMed identifier (PMID), title, abstract, and identifiers relating to data submission are retained and made available to researchers. Gene Expression Omnibus (GEO) (4) or ArrayExpress (5) identifiers are used to establish computational links between the literature and any associated data. If data is publicly available, the system accesses the repository and downloads the data files. These are then fed into the relevant analytical pipelines automatically.
Data types and source
The number of breast-related samples available from BCNTB bioinformatics has increased from 2574 to 8173. This is attributable to expansions in the data types available and the resources from which these data were obtained (Table 1).
Table 1. Overview of the analyses, data types and visualisation options available from BCNTB:Analytics.
| Features | BCNTB:Analytics | Visualisation | R Package(s) | |||
|---|---|---|---|---|---|---|
| PubMed | TCGA | CCLE | ||||
| ANALYTICAL MODULES | Novel | |||||
| PCA | ✓ | ✓ | ✓ | Scatterplots | plotly (8) | |
| Histogram | ||||||
| Correlation | ✓ | ✓ | ✓ | Heatmap | gplots (9), heatmaply (10), plotly (8) | |
| Mutation analysis | ✓ | ✓ | ✓ | Heatmap | TCGAbiolinks (11) | |
| Barplots | ||||||
| Copy number analysis | ✓ | Frequency plots | heatmaply (10), plotly (8) | |||
| Boxplots | ||||||
| Gene network visualisation | ✓ | ✓ | ✓ | Network | visNetwork (12) | |
| Data integration | ✓ | Scatterplots | plotly (8) | |||
| Boxplots | ||||||
| Enhanced | ||||||
| Tumour purity | ✓ | ✓ | Scatterplot | estimate (13) | ||
| Dynamic table | ||||||
| Molecular subtyping | ✓ | ✓ | Barplot | genefu (14) | ||
| Dynamic table | ||||||
| Molecular receptor status | ✓ | ✓ | Stacked histogram | mclust (15) | ||
| Dynamic table | ||||||
| Expression profiles | ✓ | ✓ | ✓ | Boxplot | plotly (8) | |
| Sample-level histogram | ||||||
| Survival analysis | ✓ | ✓ | Kaplan Meier plot | survival (16) | ||
| - univariate | ||||||
| - multivariate | ||||||
| Dynamic table | ||||||
| SPECIMENS | Specimen type | Tissue | Tissue | Cell lines | ||
| Cell lines | ||||||
| DATA | Type | Gene expression | Gene expression | Gene expression | ||
| Mutation | Copy number | |||||
| Mutation | ||||||
| OUTPUT | Download | Static plots (png) | Static plots (png) | Static plots (png) | ||
| Clinical & molecular data | Clinical & molecular data | Clinical & molecular data | ||||
| Graphics | Interactive plots | |||||
| Dynamic tables | ||||||
| Publication details | PMID, Title, Author, Abstract | |||||
| Hyperlink to raw data | ||||||
| Status of manual curation | ||||||
| ASSISTANCE | Documentation | Available online | ||||
| Short courses | Run throughout the year | |||||
| Help/feedback | Available online | |||||
Not only has the amount of transcriptomic data increased but users also have access to sequencing, genomic and mutational data. Originally, all data was obtained from GEO but BCNTB bioinformatics now incorporates data from major national and international cancer efforts.
PubMed (enhanced)
Adoption of SMAC, development of novel workflows and optimisation of existing workflows has allowed for a significant growth in transcriptomic data (n=5707). The analytical workflow is no longer dependent on specific annotation dictionaries. Instead, annotations are extracted from the Investigation Description Format (IDF) files available from ArrayExpress or the SOFT formatted family file(s) available from GEO. In addition, the data values are also downloaded as Series Matrix File(s) and incorporated into the relevant analytical workflows. As such, data generated from a range of platforms is available. By automating this process, we allow for a faster turnaround time for data identification, acquisition, preparation and analysis. We appreciate that a common barrier when attempting to access data programmatically is inconsistencies in the manner in which data is reported. Implementing automated processes often leads to a sacrifice of data quality for data quantity. To address this issue, all records uploaded to BCNTB:Analytics will also be manually curated by the tissue bank team. So, while datasets will be made available to the breast cancer research community from the moment they are analysed, datasets that have been reviewed are denoted by a blue star. We encourage researchers to upload their own data to the portal (http://bioinformatics.breastcancertissuebank.org/upload.jsp) and are preparing data standards to facilitate this process.
The Cancer Genome Atlas (novel)
The Cancer Genome Atlas (TCGA) (6) was developed to characterise the spectrum of genetic alterations associated with malignant transformation, including changes to the DNA sequence, copy number alterations, chromosomal aberrations and epigenetic modifications. Over 2.5 petabytes of genomic data, covering 33 different tumour types, has been generated. We aim to access and download breast cancer-specific data available from the TCGA and allow researchers to analyse these. Currently, expression (n=1205) and mutation (n=1097) data generated using RNA-seq is available for analysis alongside data from the BCNTB. Genomic and methylation data will become available in the next release.
Cancer Cell Line Encyclopedia (novel)
The Cancer Cell Line Encyclopedia (CCLE) (7) is a compilation of data generated from human cancer cell lines. Breast cancer data generated using RNA-seq, expression (n=56) and mutation (n=54), and genomic data (Affymetrix Genome-Wide Human SNP 6.0 array, n=54) has been isolated from the CCLE and is available for analysis.
BCN tissue bank (novel)
A data return policy has been implemented by the BCNTB. This specifies that data generated using BCNTB tissues must be returned to the Bank and made available to future researchers. BCNTB bioinformatics will host the returned data, publication data and sample characteristics, and make it available for analysis. As the molecular and clinical data for each patient/sample continues to increase, there will be the opportunity to conduct integrative analyses, allowing for a multidimensional understanding of the samples, their alterations and the relationships underlying them. Researchers will also have the opportunity to specify samples of interest and request matched or similar specimens.
Analytical options
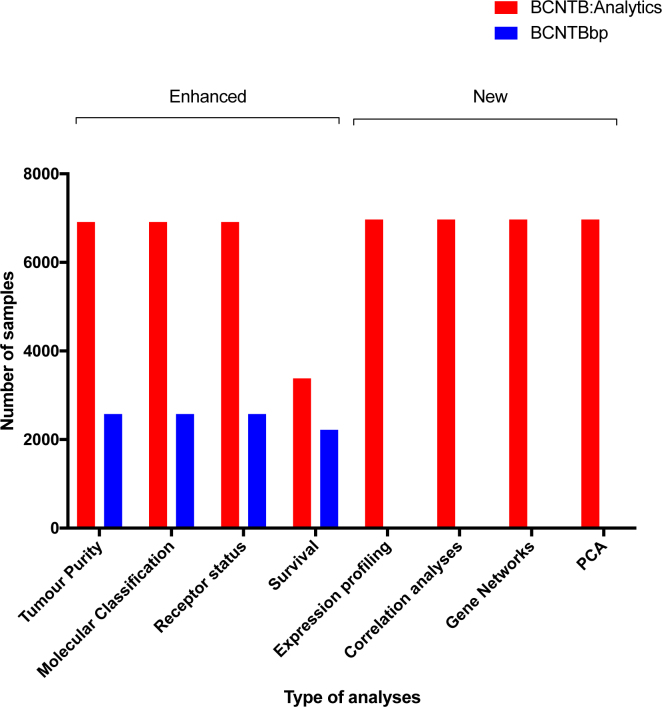
The structure of BCNTB:Analytics has been expanded and adopts a three-tiered analytical approach: exploratory, investigative and interpretative. In addition to enhancing analyses available from the previous release, we also introduce many novel features such as correlation analyses, identification of copy number aberrations, visualisation of mutational profiles, gene network analyses and an integrative modality in which data generated by different technologies is overlaid (Figure 1). Furthermore, experimental data returned to the BCNTB will be available for investigation. Any novel findings resulting from these analyses will be uploaded to the tissue bank and the associated clinical/molecular information will be updated.
Figure 1.
Growth and expansion of the data and analytical options available from BCNTB:Analytics.
Principal component analysis (new feature; Figure S1)
The inclusion of principal component analyses (PCA) (17) allows users to conduct exploratory analyses by reducing the complexity of multivariate data whilst minimising the loss of information. Data are transformed into a coordinate system and presented as an orthogonal projection. This allows researchers to visualise the global structure of the data and identify key ‘components’ of variation. For each dataset, scatterplots representing the first two and the first three principal components (PCs) are presented. To provide a better understanding of the global variability of the data, the fraction of total variance attributed to each PC is also provided.
Tumour purity (enhanced)
Cancer samples frequently contain a small proportion of infiltrating stromal and immune cells that may not only confound the tumour signal in molecular analyses but may also have a role in tumourigenesis and progression. We apply the ESTIMATE algorithm (13) that uses gene expression data to calculate stromal score, immune score and estimate score, and infers tumour purity from these values. To facilitate the understanding of these scores and their implications in inferring tumour purity we now present all the calculated scores in a single interactive scatterplot and the related values in a dynamic table that can be filtered by key word or tumour purity value.
Molecular classification (enhanced)
The analyses underlying this module remain unchanged. Each sample is assigned to a molecular subtype and a molecular receptor status for oestrogen (ER), progesterone (PR) and Her2 is calculated. The results are presented in a more comprehensive manner than in the previous release, with full annotations and sample-level information available for download.
Expression plots (enhanced with new features; Figure S2)
The code and data underlying these analyses has been optimised and expanded. Complementarity between the automated and manual process implemented by BCNTB:Analytics allows for the underlying clinical data to be more granular. In turn, these manually-curated comparative groups form the basis of the expression plots, allowing for greater insights to be obtained from the expression profiling process.
Correlation analysis (new feature; Figure S3)
The Pearson Product Moment Correlation Coefficient (PMCC) is applied to define the relationship between user-defined genes. These are presented in a heatmap in which the colour of the cell represents the correlation coefficient for each comparison. The calculated correlation coefficient can be viewed for each pairwise comparison by hovering over the heatmap.
Survival analysis (enhanced)
In addition to the summary of survival, based on molecular subtype and a univariate (single gene) model, available from the previous version, the effect of multiple genes can also be investigated. Researchers can either conduct the analyses on all samples in a dataset or choose to focus on a subtype(s) of interest. Results are presented as Kaplan Meier plots. A summary of survival covariates (P-value and hazard ratio) for five-, ten- and overall survival are presented in tabular format.
Copy number aberrations (new feature)
A landscape of copy number alterations can be viewed at both a region- and a gene-level. An overview of copy number aberrations, specific to each biological group in the dataset, are presented as frequency plots. Heatmaps displaying the copy number status of a region in which user-defined gene(s) reside can also be generated.
Mutational profiles (new feature; Figure S4)
A summary of the top somatic single nucleotide variants from matched tumour-normal samples, as called by MuTect2 (18), can be viewed as a heatmap. Sample information, such as the number of mutations per sample and clinical features, and gene level information, such as the types of mutations identified per gene, are also presented. In addition to filtering results by top mutations, researchers also have the ability to investigate the mutational status of genes of interest.
Gene network analysis (new feature; Figure S5)
For each comparative group defined in the dataset, the interactions between genes of interest and their primary neighbours can be displayed in an interactive network. This new feature takes advantage of the mentha interactome browser (19), which collects manually-curated interactions from databases that have adhered to the IMEx consortium (20). In these gene networks, nodes represent the genes while edges represent the interactions. Nodes are coloured according to the expression level (z-score) in the dataset of interest. A detailed report of the interactions composing the network, together with the list of PMIDs that support each relationship, is provided in tabular format.
Integration of multidimensional data (new feature)
Providing researchers with the means to overlay layers of molecular information to gain a multidimensional view of the data allows for the identification of alterations that co-exist within a sample and helps provide greater insight into the relationships between them.
Data are processed using a standardised workflow to ensure comparability, reusability and interoperability both across different datasets and different data types. This ensures compatibility between a dataset(s) in which the same specimen has been analysed using different approaches, allowing for the merging of data obtained from different technologies. This analytical module allows researchers to integrate and visualise discrete genetic events, such as DNA copy-number alterations (CNAs) and mutations, or relative linear copy-number values with continuous mRNA abundance data for user-defined gene(s). For example, overexpression of a group of genes could indicate synergy between these genes or could be attributable to copy number alterations.
Data return and data sharing
BCNTB bioinformatics has been designed to allow data sharing, discoverability and re-usability. This platform is not a bioinformatics silo but rather a niche within the BCN tissue banking ecosystem that offers an unparalleled opportunity to add informative layers of molecular data to existing patient data available from the Bank.
From the BCN Tissue Bank tab, researchers will be able to view a summary of all projects using samples from the BCNTB, along with an indication about publications. Once molecular data is returned to the Bank and integrated into platform, researchers will be able to analyse the data (BCNTB:Analytics), query the published findings (BCNTB:Miner), link to the raw data files, and request specimens from the BCNTB on matched patients.
Interactive graphics and tables
All results retrieved by the portal are presented in an interactive format (8). The nature of these results allows researchers to zoom in/out, focus on areas of interest, visualise the annotation of data points, and exclude or include samples in pre-defined biological groups. Where applicable, information and results are also presented in tabular format. These are also dynamic, allowing samples to be filtered by features of interest. All figures can be downloaded as static files of publishing quality.
For each dataset, the molecular results from which the figures are generated, such as those from the molecular subtyping of samples and tumour purity estimates, are presented in an interactive table. This allows for results to be inspected at a sample level. These results can be copied or downloaded in the most common formats (excel spreadsheet, csv or pdf) for subsequent analyses.
BCNTB: Miner
Restructuring of the data-mining architecture
BCNTB:Miner has migrated to a more powerful server, improving response time to queries. During this time, two major restructuring processes were completed—the core architecture of the database was revised and the annotation/mappings systems were updated. The restructured data-mining modality encompasses a user-friendly interface with a clean design that emphasises the querying parameters available.
Query building and annotation categories
The data management system underlying the data-mining module was updated to BioMart 0.9 (21). This has allowed for information from the Study Dataset and Gene Dataset, available in the previous release, to be merged and integrated with Ensembl features, such as genes, transcripts, proteins, SNPs, genomic features, gene ontologies, multi-species comparisons, and variant source.
The request-response architecture remains the same. However, the novel environment offered by BioMart 0.9, in conjunction with the MartWizard GUI adopted by BCNTB:Miner, allows for the querying process to be more instinctual than ever before. This interface guides the user through a query in a logical manner and presents all the parameters selected in an additive manner on the right-hand side of the page. The simplification of the interface is accompanied by optimisation in the structure of the underlying MySQL tables. Moreover, updating the annotations and mappings from Ensembl 69 to Ensembl 84 ensures that results retrieved are mapped using up-to-date annotations.
All of these changes help ensure that researchers continue to experience improvements in the querying process without having to compromise on the complexity or granularity of the query. BCNTB:Miner provides users with an intuitive interface to build custom queries against all the genomics, methylomics, transcriptomics, proteomics and microRNA data that has been mined from the literature and linking to pathways and mechanisms involved in breast cancer.
DOCUMENTATION AND AVAILABILITY OF THE PLATFORM
A comprehensive user guide is available from the homepage. The tissue bank bioinformatics team are also available to answer any additional queries at bioinformatics.breastcancertissuebank@qmul.ac.uk. Furthermore, we host BCN-funded ‘breast cancer bioinformatics’ courses in which BCNTB bioinformatics is showcased and explored in depth.
BCNTB bioinformatics was, and continues to be, designed and developed for researchers. As such, we welcome any feedback, suggestions or ideas regarding how to improve our resources. An expandable Feedback tab is available for this purpose.
BCNTB bioinformatics is freely available and has been tested using Google Chrome, Mozilla Firefox and Safari on Mac OS X10.12.4 and Windows OS.
LOOKING TO THE FUTURE
The unique relationship between BCNTB bioinformatics and the Tissue Bank has phenomenal potential in ensuring that breast cancer data is exploited to its maximal potential. Not only will we integrate and update specimen/patient data that is returned to the BCNTB but researchers will also be able to analyse and integrate their own data into the portal. To help facilitate this process, we are preparing data standards for data generated on different platforms. The standardisation of the data content and structure, in conjunction with a manual assurance stage, will ensure that data can be uploaded, analysed and interpreted with ease.
In addition to increasing the amount of data available to researchers and including data from other national/international cancer initiatives, such as the International Cancer Genome Consortium (ICGC), future expansions of BCNTB bioinformatics will provide researchers with the ability to apply predictive models to the BCNTB returned data. By having access to a rich collection of clinical data and creating virtual patient models, individual tumours could be characterised at the molecular level, making it possible to develop prognostic signatures and identify the potential responses to treatment/drug repurposing.
It is vital that BCNTB bioinformatics continues to evolve alongside its in vivo and in vitro components. In future, we hope that the BCNTB and the BCNTB bioinformatics will no longer be viewed as separate entities but as niches within a biobanking research ecosystem, one in which patient benefit is core.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Breast Cancer Now (BCN) for funding this work and all the members of the BCNTB teams across the five dedicated centres. These are led by Prof Claude Chelala (Barts-bioinformatics); Prof. Louise Jones (Barts); Prof Ian Ellis and Dr Matharoo-Ball Balwir (Nottingham); Prof Angie Cox (Sheffield); Prof. Val Speirs (Leeds); and Dr Ramsey Cutress (Southampton). We would also like to thank BCNTB IT Lead John Watts, BCN staff members, patient advocates and the patients that have donated tissue, without whom this would not be possible.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Breast Cancer Campaign [TB2016BIF]; Pancreatic Cancer Research Fund (PCRFTB) [Tissue Bank grant, to J.M and A.Z.D.U.]. Funding for open access charge: Breast Cancer Campaign [TB2016BIF].
Conflict of interest statement. None declared.
REFERENCES
- 1. Thompson A., Brennan K., Cox A., Gee J., Harcourt D., Harris A., Harvie M., Holen I., Howell A., Nicholson R. et al. Evaluation of the current knowledge limitations in breast cancer research: a gap analysis. Breast Cancer Res. 2008; 10:R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cutts R.J., Guerra-Assuncao J.A., Gadaleta E., Dayem Ullah A.Z., Chelala C.. BCCTBbp: the Breast Cancer Campaign Tissue Bank bioinformatics portal. Nucleic Acids Res. 2015; 43:D831–D836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kolde R., Laur S., Adler P., Vilo J.. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012; 28:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clough E., Barrett T.. The gene expression omnibus database. Methods Mol. Biol. 2016; 1418:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y.A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T. et al. ArrayExpress update–simplifying data submissions. Nucleic Acids Res. 2015; 43:D1113–D1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cancer Genome Atlas Research, N. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M.. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sievert C., Parmer C., Hocking T., Chamberlain S., Ram K., Corvellec M., Despouy P.. Plotly Technologies Inc. 2017.
- 9. Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S.. gplots: Various R programming tools for plotting data. R package version. 2009; 2:1. [Google Scholar]
- 10. Galili T. heatmaply: Interactive Heat Maps Using’plotly’.R package version 0.6.0. 2016.
- 11. Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016; 44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Almende B., Thieurmel B.. visNetwork: Network Visualization using’vis. js’ Library. R package version 0.2. 2016; 1: [Google Scholar]
- 13. Yoshihara K., Shahmoradgoli M., Martinez E., Vegesna R., Kim H., Torres-Garcia W., Trevino V., Shen H., Laird P.W., Levine D.A. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013; 4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deena M. Computation of Gene Expression-Based Signatures in Breast Cancer. 2015. [DOI] [PMC free article] [PubMed]
- 15. Scrucca L., Fop M., Murphy T.B., Raftery A.E.. mclust 5: Clustering, classification and density estimation using gaussian finite mixture models. R J. 2016; 8:289. [PMC free article] [PubMed] [Google Scholar]
- 16. Therneau T.M., Grambsch P.M.. Modeling Survival Data: Extending the Cox Model. 2000; NY: Springer. [Google Scholar]
- 17. Jolliffe I.Y. Principal Component Analysis. 2002; 2nd edn.NY: Springer-Verlag. [Google Scholar]
- 18. Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G.. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013; 31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calderone A., Castagnoli L., Cesareni G.. mentha: a resource for browsing integrated protein-interaction networks. Nat. Methods. 2013; 10:690–691. [DOI] [PubMed] [Google Scholar]
- 20. Orchard S., Kerrien S., Abbani S., Aranda B., Bhate J., Bidwell S., Bridge A., Briganti L., Brinkman F.S., Cesareni G. et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods. 2012; 9:345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smedley D., Haider S., Durinck S., Pandini L., Provero P., Allen J., Arnaiz O., Awedh M.H., Baldock R., Barbiera G. et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015; 43:W589–W598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.