Abstract
Biological networks provide new opportunities for understanding the cellular biology in both health and disease states. We generated tissue specific integrated networks (INs) for liver, muscle and adipose tissues by integrating metabolic, regulatory and protein-protein interaction networks. We also generated human co-expression networks (CNs) for 46 normal tissues and 17 cancers to explore the functional relationships between genes as well as their relationships with biological functions, and investigate the overlap between functional and physical interactions provided by CNs and INs, respectively. These networks can be employed in the analysis of omics data, provide detailed insight into disease mechanisms by identifying the key biological components and eventually can be used in the development of efficient treatment strategies. Moreover, comparative analysis of the networks may allow for the identification of tissue-specific targets that can be used in the development of drugs with the minimum toxic effect to other human tissues. These context-specific INs and CNs are presented in an interactive website http://inetmodels.com without any limitation.
INTRODUCTION
Systems biology is an emerging tool that uses both integrative and systems-level approaches for organizing and interpreting highly complex and heterogeneous information generated by high-throughput technologies (1). Systems biology/medicine has provided a new perspective in biology and medicine and allowed for detailed understanding of how biological systems function in both health and disease states through the use of biological networks including GEnome-scale metabolic Models (GEMs), transcriptional regulatory networks (RNs), protein–protein interaction networks (PPINs), signalling networks (SNs) and co-expression networks (CNs) (2,3). To date, a large number of GEMs has been generated for major human tissues (4–8), tumor samples (9–11) as well as individual patients (12–14). Similarly, generic and tissue-specific PPINs (15,16), RNs (17,18) and SNs (19,20) have been generated and employed in understanding the relationship between genes and their key role in various biological functions in different clinical conditions. Those biological networks have provided a comprehensive perspective on the biological functions of the given tissue/cancer and used as comprehensive resources in the analysis and integration of multiple omics data. These models have been deposited to resources such as Human Metabolic Atlas (21), BIGG database (22) and BioModels (23).
Considering the interplay between the proteins and their major role in a specific biological function, we generated tissue-specific Integrated Networks (INs) by merging GEMs, RNs and PPINs for liver, adipose and muscle tissues (24). Even though INs provide detailed information about the physical link between genes, metabolites and transcription factors, these physical links do not have to result in functional relationship. In this context, we also generated tissue-specific CNs for 46 major human tissues and tested an hypothesis about the association of fatty acid synthase (FASN), the key enzyme in de novo lipid synthesis, with the progression of non-alcoholic fatty liver disease (NAFLD) and hepatocellular carcinoma (HCC) (25). Here, we presented INs for liver, muscle and adipose tissues and CNs for 46 human tissues and 17 cancers to the research community. We created a database for tissue/cancer-specific biological networks (TCSBN) and presented these networks in an interactive website http://inetmodels.com without any limitation. Our database allows users to explore the relationships between genes and biological functions in a given tissue/cancer through the employment of CNs in gene-centric manner. Each gene is linked to Human Protein Atlas (HPA) (http://www.proteinatlas.org/), where the protein and mRNA expression in major human tissues (8) and the subcellular localization of the proteins (26) are presented. Moreover, users can investigate the overlap between functional and physical interactions through the employment of INs and CNs for human tissues. We finally presented a case study where we employed these biological networks for testing a biological hypothesis that can be used in the discovery of tissue-specific drug targets for the development of efficient treatment methods.
DATABASE DESCRIPTION
We generated INs for liver, muscle and adipose tissues by integrating GEMs, PPINs and RNs (24). We used tissue-specific proteomics and transcriptomics data presented in HPA (8) to refine the generic human PPIN (27) and RN that is generated using ENCODE DNase-seq data (28). INs have been employed in the analysis of the transcriptomics data obtained from lean and obese subjects and deregulation around mannose metabolism has been elucidated (24). The study proposed that plasma mannose levels can be used in prediction of the insulin resistance independently of BMI. In a follow up study, plasma mannose levels have been analyzed with specific liquid chromatography- mass spectrometry in relation to future development (up to 8 years) of Type 2 diabetes (T2D), cardiovascular disease (CVD) and diabetic kidney disease in a total of over 8,000 individuals who did not have disease at the time of the baseline mannose levels (29). We found that elevated plasma mannose levels are strong markers of future risk of several chronic diseases and that it may contribute to their development rather than just being a novel biomarker (29). It has been demonstrated that tissue-specific INs are valuable tools in the analysis of clinical data and provide useful information that can be translated to the clinic.
Moreover, we generated CNs for 46 human tissues and 17 cancers using the tissue and cancer specific RNA-seq data presented in GTEx (30) and TCGA (31) databases, respectively. RNA-seq data of those databases were processed with different pipelines and counted with different units, such as transcripts per million (TPM) for human tissues in GTEX (30) and fragments per kilobase of transcript per million mapped reads (FPKM) for cancers in TCGA (31). Therefore, to minimize the biases during the generation of CNs, we excluded lowly-expressed genes based on their median gene expression (less than 1 TPM for human tissues; less than 1 FPKM for cancers) and combined all top-100 (i.e. positive correlation) and bottom-100 (i.e. negative correlation) co-expressed genes of remaining genes on the networks, based on Pearson's correlation coefficients. Of note, the number of genes included in each network may vary since the mRNA expression of a gene is different in each tissue or cancer. Those human tissue networks included the smallest network of whole blood (10 063 nodes) and the largest network of testis (15 621 nodes) and human cancer networks included the smallest network of liver cancer (10 037 nodes) and the largest network of glioma (12 147 nodes) (Supplementary Table S1).
We identified liver-specific targets that can be used in the development of drugs for liver diseases with the minimum toxic effect to other human tissues. Moreover, we generated cancer-specific co-expression networks of 17 major human cancers (32). We found that a large fraction of genes is differentially expressed in cancers and these genes have an impact on overall patient survival. We also showed that gene expression patterns of individual tumours varied considerably, and could exceed the variation observed between different cancer types. Shorter patient survival was generally associated with up-regulation of genes involved in mitosis and cell growth, and down-regulation of genes involved in cellular differentiation. We also found that genes involved in hallmarks of cancer (33,34) were mostly co-expressed with prognostic genes in each cancer. For all cancers, we found co-expression clusters enriched with prognostic genes and hallmark genes and identified potential targets that may be used in the development of anti-cancer drugs.
NETWORK SEARCH
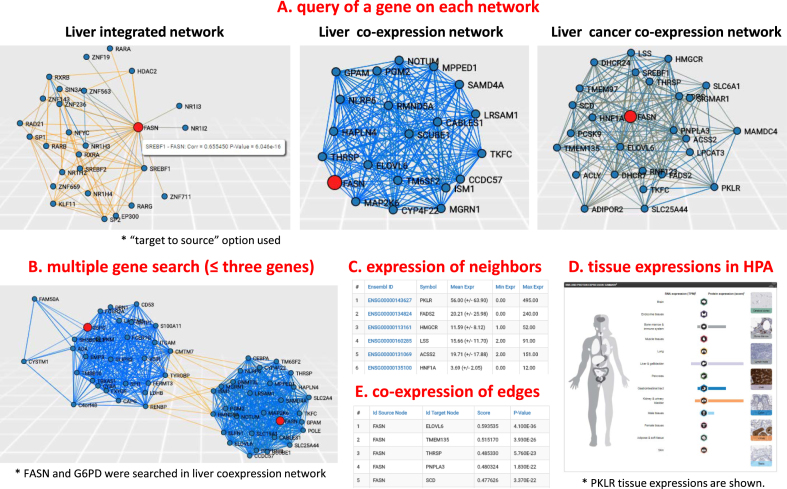
Users may explore neighbours of a query gene in each biological network (Figure 1) to identify the genes that either directly interacts or co-expresses with the queried gene. Choosing a query gene in a network type, users may select the direction to find neighbours of a query gene: from source node to target node, or vice versa. For integrated networks, this option is useful in finding the regulators of a query gene (by ‘target to source’ option) or target genes (by ‘source to target’ option). In Figure 1A, we used ‘target to source’ option to find regulators of FASN in liver integrated network. In addition, the user may set a parameter to describe a maximal number of nodes for neighbours of a queried gene to limit the network size for visualization purposes. We ranked the neighbour genes based on the absolute correlation coefficients and showed them with corresponding correlation coefficients and P-values. Therefore, users can choose neighbours of a query gene with desired statistical significances. The database includes another parameter, called edge pruning parameter, to show only edges with desired statistical significances. We set default P-value as 0.01, but users can change it in a full range of P-values.
Figure 1.
Description of the database to explore the neighbours of a query gene. (A) Users can input a query gene in a network type to explore and constrain a network size to visualize by setting maximum number of neighbour nodes. For instance, we queried fatty acid synthase (FASN) in liver integrated network, liver co-expression network and liver cancer co-expression network. Of note, ‘target to source’ option was used to find regulators of FASN in liver integrated network. (B) The users are allowed for searching multiple genes (up to three genes). We showed an example for searching FASN and G6PD simultaneously in liver co-expression network and found their neighbours are independently clustered. (C) Along with a visualized network from a query gene, it also shows tables of a query gene and neighbours with their expression values of given tissue. (D) Through hyperlinks of each gene to Human Protein Atlas (HPA), users can explore expression in human tissues and cancers as well as the subcellular localization of the gene. The expression of PKLR in HPA showed its tissue-specific expressions, especially in liver and kidney tissues. (E) We showed correlation coefficients and P-values of visualized edges, enabling users to check statistical significance of the searched network.
As an example, we searched a transporter involved in the transport of fatty acids from cytosol to mitochondria, carnitine palmitoyltransferase I (CPT1A) in the liver co-expression network (Supplementary Figure S1) by setting maximum number of nodes to 25 and edge pruning parameter to 2 (i.e. –log 10P = 2.0). We identified three clusters of co-expressed neighbours (red circles, Supplementary Figure S1A). Interestingly, a cluster that is negatively correlated to CPT1A includes enzymes involved in lipid synthesis. We also found two other clusters that include genes involved in the beta-oxidation of fatty acids, electron chain transport, lipid transport and mitochondrial biogenesis, which are associated with the function of CPT1A. Likewise, users can search their genes of interest and identify functionally related genes in a given tissue or cancer.
The users are allowed for searching multiple genes (as queries up to three genes) that may help to see the co-expression landscape of genes of interest, simultaneously (Figure 1B and Supplementary Figure S1B). For example, we searched CPT1A and FASN, which have different functions in lipid metabolism and found to be independently clustered and negatively correlated in liver co-expression networks (Supplementary Figure S1A). In addition, we checked FASN and glucose-6-phosphate dehydrogenase (G6PD), which are enzymes inversely metabolizing NADPH, and identified independent clusters that are negatively correlated in liver co-expression network (Figure 1B). Likewise, users may check genes of interest if they are clustered together in a given tissue or cancer network.
We also showed expressions of genes in selected tissues at ‘Search Genes’ and ‘Related Genes’ tables: mean with standard deviation, minimum and maximum expressions in each tissue sample included in the analysis. For INs, we used gene expression information from GTEx RNA-seq of corresponding tissues. Hyperlinks of the shown genes enable users to check their protein expressions in HPA (8). For example, when we checked neighbours of CPT1A by the hyperlinks (Supplementary Figure S1A), we found Apolipoprotein A-V (APOA5) as liver-tissue enriched gene in HPA database, whereas CPT2 as expressed in all human tissues. Thus, the user can check the tissue-specificity of a gene through hyperlinks to HPA. We also showed co-expression values and P-values on edges of visualized networks as ‘Edges’ tables for all types of networks (Figure 1E). All network figures and expression as well as edge tables can be downloaded as image file and CSV file, respectively. Edge tables saved as CSV file can be imported into Cytoscape (www.cytoscape.org) for more detailed network visualization and modularization (see tutorial page).
A CASE STUDY RELATED TO FASN
A major strength of our database is to explore co-expression landscape over various human tissues and cancers. In our recent work, we found that co-expression gene clusters were associated with tissue-specific functions and enabled the testing of biological hypotheses in a given tissue context (25). Notably, tissue-specific CNs allowed for the identification of tissue-specific drug targets with minimum toxic effects expected for other tissues. Our recent study also showed that co-expression gene clusters might assist in identifying cancer driver genes when it is combined with clinical metadata (32). Here, we presented a case study to explore co-expression landscape for testing a biological hypothesis, which may benefit research community.
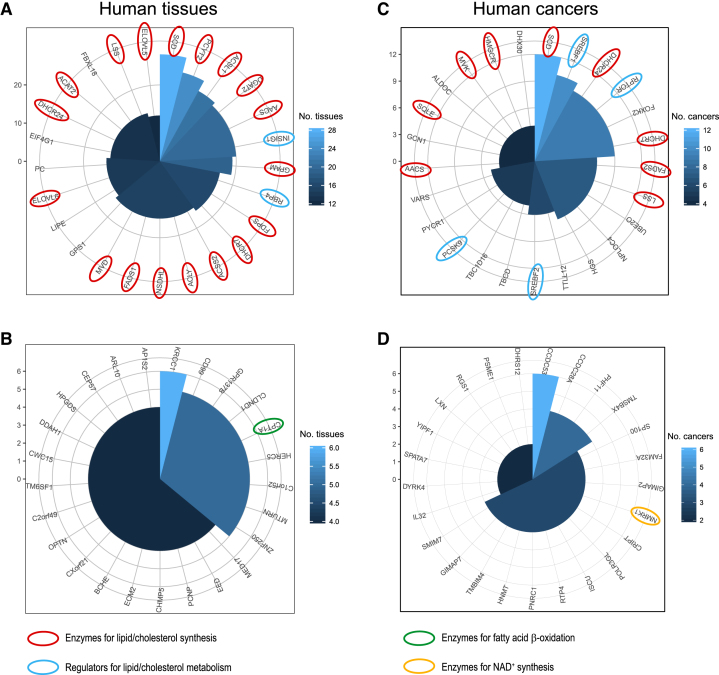
Lipids serve as essential substrates for cell membrane, signalling and energy storage and their synthesis is strictly regulated in a cell and catalysed by a key enzyme, FASN (35,36). Based on co-expression landscapes of over 46 human tissues, we studied how FASN is regulated together with other genes. Among top-100 co-expressed (i.e. positive correlation) genes of FASN in all human tissues, we studied the 25 genes that are most frequently identified among human tissues (Figure 2A). Intriguingly, we found that the majority of them are involved in lipid/cholesterol metabolism: eighteen genes (72%) were enzymes and involved in lipid/cholesterol synthesis and two genes (8%) were regulators of lipid/cholesterol metabolism. Based on DAVID gene ontology enrichment analysis (https://david.ncifcrf.gov/), we found that these genes are significantly enriched in lipid metabolic process and cholesterol biosynthetic process (FDR 2.19 × 10−18 and 3.68 × 10−10, respectively). Notably, the most co-expressed gene in all human tissues was stearoyl-CoA desaturase-1 (SCD), which catalyses the conversion of stearoyl-CoA to oleoyl-CoA, a subsequent step of fatty acid synthesis by FASN. In addition, among the 25 most co-expressed genes were the enzymes, pyruvate carboxylase (PC) and intracellular lipase (LIPE), which are highly relevant to fatty acid synthesis from pyruvate and dietary lipids, respectively. In contrast, we found that the bottom-100 co-expressed (i.e. negative correlation) genes with FASN in all human tissues were rarely associated with fatty acid synthesis (Figure 2B). Rather, one of most-negatively correlated genes to FASN in all tissues was CPT1A.
Figure 2.
A comparative analysis of co-expression landscape over 46 human tissues and 17 cancers. Among top-100 coexpressed genes of 46 human tissues or 17 cancers, we selected those genes that are shown mostly in tissues (A) or cancers (C), respectively. Polar coordinate indicates how many tissues or cancers they were shown as top-co-expressed genes. Likewise among bottom-100 co-expressed genes of 46 human tissues or 17 cancers, we selected those that are shown mostly in tissues (B) or cancers (D), respectively.
Lipid synthesis is required for cellular proliferation and FASN is considered to be a therapeutic target for cancer treatment (37,38). Therefore, we analysed how FASN is regulated in cancers, based on co-expression landscape of FASN over 17 cancers (Figure 2C): among the top-100 co-expressed genes in all cancers, we selected the top-25 genes that are most frequently shown among cancers. Even though cancers are known to show high heterogeneity, we found that more than half of top-25 genes in all cancers were associated with lipid metabolism: nine genes (36%) were enzymes for lipid/cholesterol synthesis and four genes (16%) were regulators for lipid/cholesterol metabolism. Based on DAVID analysis, we also found that they were significantly enriched in lipid metabolic process and cholesterol metabolic process (FDR 4.06 × 10−9 and 0.00112, respectively). We also studied the bottom-100 co-expressed genes in all cancers, as we analysed in human tissues (Figure 2D). We observed that they were rarely associated with FASN. Rather, one of them was associated with NAD+ synthesis, which is required for fatty acid beta oxidation. In addition, our analysis of tissue-specific targets based co-expression landscape, yielded similar observations reported in our previous work (25) (Figure 1). For example, we can check neighbours of FASN in liver cancer co-expression network and also their expressions in normal tissues from hyperlinks to HPA.
Among the top-100 co-expressed genes of FASN, most genes were frequently found in all human tissues, showing less tissue-specificity. From HPA, the users can find the tissue-specificity of a given gene and identify tissue-specific drug targets. For example, PKLR, the 10th top-coexpressed gene with FASN in the liver cancer co-expression network, was found to be liver tissue-specific based on HPA and it has been identified as a drug target (Figure 1C and D). Furthermore, an integrated tissue network may assist in identifying the regulators of given gene targets thus enabling to investigate mechanistic details (Figure 1A).
CONCLUSION
We generated tissue and cancer specific biological networks and presented them in a freely accessible interactive website http://inetmodels.com. We showed that our resource may benefit the research community to test a hypothesis and identify potential tissue-specific drug targets that may be used in the development of efficient treatment strategies. The case study presented about FASN demonstrated the successful use of our networks in testing a biological hypothesis. We found that the co-expression landscape provided known biological knowledge, such as regulation of lipid/cholesterol metabolism, important for cancer. In addition, we identified tissue-specific drug targets, expecting less toxic effects on other human tissues, and also regulators of given targets, based on physical interactions provided by INs. Hence, we envisage that our database is a useful resource for the analysis of clinical data and for revealing the underlying molecular mechanisms involved in the progression of the complex diseases as previously exemplified (39–42).
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Knut and Alice Wallenberg Foundation. Funding for open access charge: Knut and Alice Wallenberg Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mardinoglu A., Nielsen J.. Systems medicine and metabolic modelling. J. Intern. Med. 2012; 271:142–154. [DOI] [PubMed] [Google Scholar]
- 2. Mardinoglu A., Gatto F., Nielsen J.. Genome-scale modeling of human metabolism - a systems biology approach. Biotechnol. J. 2013; 8:985–996. [DOI] [PubMed] [Google Scholar]
- 3. Mardinoglu A., Nielsen J.. New paradigms for metabolic modeling of human cells. Curr. Opin. Biotech. 2015; 34:91–97. [DOI] [PubMed] [Google Scholar]
- 4. Mardinoglu A., Agren R., Kampf C., Asplund A., Uhlen M., Nielsen J.. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014; 5:3083. [DOI] [PubMed] [Google Scholar]
- 5. Mardinoglu A., Agren R., Kampf C., Asplund A., Nookaew I., Jacobson P., Walley A.J., Froguel P., Carlsson L.M., Uhlen M. et al. Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Mol. Syst. Biol. 2013; 9:649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Varemo L., Scheele C., Broholm C., Mardinoglu A., Kampf C., Asplund A., Nookaew I., Uhlen M., Pedersen B.K., Nielsen J.. Proteome- and transcriptome-driven reconstruction of the human myocyte metabolic network and its use for identification of markers for diabetes. Cell Rep. 2015; 11:921–933. [DOI] [PubMed] [Google Scholar]
- 7. Bordbar A., Feist A.M., Usaite-Black R., Woodcock J., Palsson B.O., Famili I.. A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology. BMC Syst. Biol. 2011; 5:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 9. Yizhak K., Chaneton B., Gottlieb E., Ruppin E.. Modeling cancer metabolism on a genome scale. Mol. Syst. Biol. 2015; 11:817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Agren R., Bordel S., Mardinoglu A., Pornputtapong N., Nookaew I., Nielsen J.. Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput. Biol. 2012; 8:e1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Björnson E., Mukhopadhyay B., Asplund A., Pristovsek N., Cinar R., Romeo S., Uhlen M., Kunos G., Nielsen J., Mardinoglu A.. Stratification of hepatocellular carcinoma patients based on acetate utilization. Cell Rep. 2015; 13:2014–2026. [DOI] [PubMed] [Google Scholar]
- 12. Agren R., Mardinoglu A., Asplund A., Kampf C., Uhlen M., Nielsen J.. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol. Syst. Biol. 2014; 10:721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mardinoglu A., Bjornson E., Zhang C., Klevstig M., Soderlund S., Stahlman M., Adiels M., Hakkarainen A., Lundbom N., Kilicarslan M. et al. Personal model-assisted identification of NAD+ and glutathione metabolism as intervention target in NAFLD. Mol. Syst. Biol. 2017; 13:916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yizhak K., Gaude E., Le Devedec S., Waldman Y.Y., Stein G.Y., van de Water B., Frezza C., Ruppin E.. Phenotype-based cell-specific metabolic modeling reveals metabolic liabilities of cancer. eLife. 2014; 3:e03641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bossi A., Lehner B.. Tissue specificity and the human protein interaction network. Mol. Syst. Biol. 2009; 5:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Las Rivas J., Fontanillo C.. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 2010; 6:e1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Neph S., Stergachis A.B., Reynolds A., Sandstrom R., Borenstein E., Stamatoyannopoulos J.A.. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012; 150:1274–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pechenick D.A., Payne J.L., Moore J.H.. Phenotypic robustness and the assortativity signature of human transcription factor networks. PLoS Comput. Biol. 2014; 10:e1003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Helikar T., Konvalina J., Heidel J., Rogers J.A.. Emergent decision-making in biological signal transduction networks. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee S., Mardinoglu A., Lee D., Nielsen J.. Dysregulated signaling hubs of liver lipid metabolism reveal hepatocellular carcinoma pathogenesis. Nucleic Acids Res. 2016; 44:5529–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pornputtapong N., Nookaew I., Nielsen J.. Human metabolic atlas: an online resource for human metabolism. Database. 2015; 2015:bav068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. King Z.A., Lu J., Drager A., Miller P., Federowicz S., Lerman J.A., Ebrahim A., Palsson B.O., Lewis N.E.. BiGG Models: a platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016; 44:D515–D522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chelliah V., Juty N., Ajmera I., Ali R., Dumousseau M., Glont M., Hucka M., Jalowicki G., Keating S., Knight-Schrijver V. et al. BioModels: ten-year anniversary. Nucleic Acids Res. 2015; 43:D542–D548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee S., Zhang C., Kilicarslan M., Piening B.D., Bjornson E., Hallstrom B.M., Groen A.K., Ferrannini E., Laakso M., Snyder M. et al. Integrated network analysis reveals an association between plasma mannose levels and insulin resistance. Cell Metab. 2016; 24:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee S., Zhang C., Uhlen M., Nielsen J., Smith U., Boren J., Mardinoglu A.. Network analyses identify liver-specific targets for treating liver diseases. Mol. Syst. Biol. 2017; 13:938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L.M. et al. A subcellular map of the human proteome. Science. 2017; 356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 27. Rolland T., Tasan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R. et al. A proteome-scale map of the human interactome network. Cell. 2014; 159:1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mardinoglu A., Stancakova A., Lotta L.A., Kuusisto J., Boren J., Bluher M., Wareham N.J., Ferrannini E., Groop P.H., Laakso M. et al. Plasma mannose levels are associated with incident type 2 diabetes and cardiovascular disease. Cell Metab. 2017; 26:281–283. [DOI] [PubMed] [Google Scholar]
- 30. Consortium G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cancer Genome Atlas Research, N. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M.. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Uhlen M., Zhang C., Lee S., Sjostedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F. et al. A pathology atlas of the human cancer transcriptome. Science. 2017; 357:eaan2507. [DOI] [PubMed] [Google Scholar]
- 33. Hanahan D., Weinberg R.A.. The hallmarks of cancer. Cell. 2000; 100:57–70. [DOI] [PubMed] [Google Scholar]
- 34. Hanahan D., Weinberg R.A.. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 35. Wakil S.J., Stoops J.K., Joshi V.C.. Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 1983; 52:537–579. [DOI] [PubMed] [Google Scholar]
- 36. Pandey P.R., Liu W., Xing F., Fukuda K., Watabe K.. Anti-cancer drugs targeting fatty acid synthase (FAS). Rec. Pat. Anticancer Drug Discov. 2012; 7:185–197. [DOI] [PubMed] [Google Scholar]
- 37. Currie E., Schulze A., Zechner R., Walther T.C., Farese R.V. Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013; 18:153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Menendez J.A., Lupu R.. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 2007; 7:763–777. [DOI] [PubMed] [Google Scholar]
- 39. Bosley J., Boren C., Lee S., Grotli M., Nielsen J., Uhlen M., Boren J., Mardinoglu A.. Improving the economics of NASH/NAFLD treatment through the use of systems biology. Drug Discov. Today. 2017; 22:1532–1538. [DOI] [PubMed] [Google Scholar]
- 40. Benfeitas R., Uhlen M., Nielsen J., Mardinoglu A.. New challenges to study heterogeneity in cancer redox metabolism. Front Cell Dev. Biol. 2017; 5:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nielsen J. Systems biology of metabolism: a driver for developing personalized and precision medicine. Cell Metab. 2017; 25:572–579. [DOI] [PubMed] [Google Scholar]
- 42. Bjornson E., Boren J., Mardinoglu A.. Personalized cardiovascular disease prediction and treatment—a review of existing strategies and novel systems medicine tools. Front. Physiol. 2016; 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.