Abstract
ProteomicsDB (https://www.ProteomicsDB.org) is a protein-centric in-memory database for the exploration of large collections of quantitative mass spectrometry-based proteomics data. ProteomicsDB was first released in 2014 to enable the interactive exploration of the first draft of the human proteome. To date, it contains quantitative data from 78 projects totalling over 19k LC–MS/MS experiments. A standardized analysis pipeline enables comparisons between multiple datasets to facilitate the exploration of protein expression across hundreds of tissues, body fluids and cell lines. We recently extended the data model to enable the storage and integrated visualization of other quantitative omics data. This includes transcriptomics data from e.g. NCBI GEO, protein–protein interaction information from STRING, functional annotations from KEGG, drug-sensitivity/selectivity data from several public sources and reference mass spectra from the ProteomeTools project. The extended functionality transforms ProteomicsDB into a multi-purpose resource connecting quantification and meta-data for each protein. The rich user interface helps researchers to navigate all data sources in either a protein-centric or multi-protein-centric manner. Several options are available to download data manually, while our application programming interface enables accessing quantitative data systematically.
INTRODUCTION
Mass spectrometry has developed into the flagship technology for proteome research much akin to what next generation sequencing has become for genomics and transcriptomics (1,2). Since proteins execute and control most biological processes in all domains of life, they are one of the most frequently targeted class of molecules in the context of drug development. Today, scientists and clinicians anticipate that proteins will also become a major source of biomarkers (3) useful to diagnose disease, to stratify patients for treatment and to monitor response to therapy to name a few.
At the same time, the volume and complexity of proteomics data generated by modern mass spectrometers is challenging our ability to turn data into tractable hypotheses, within and, particularly, across larger projects. In order to provide access to previously performed experiments, many different repositories have been developed (4,5). However, their focus is often limited to a particular aspect of the data and frequently, protein identification is de-coupled from protein quantification. PRIDE (6) is currently the community-standard for publishing raw data but also peptide and protein identification results (including post-translational modifications). However—until recently—it lacked an intuitive interface for comparing results across different datasets. PeptideAtlas (7), GPMDB (8) and MASSIVE mostly focus on hosting identification results by re-processing data using their own pipelines. The protein abundance database (PAXDB) (9) stores quantification data from publicly available data, but lacks the underlying peptide identification results. MaxQB (10) does provide both protein identification and quantification data, but is far less comprehensive than any of the other repositories and also does not allow cross-dataset comparison. While most of these databases can store meta-data such as sample preparation and data acquisition protocols, specific treatments and the different conditions used in the experimental setup are not stored in a programmatically accessible format. In addition, none of the aforementioned databases allow storage of other data types. This in turn makes it difficult to systematically explore and mine data across proteomic or multi-omics experiments.
ProteomicsDB is filling this gap by not only enabling cross-dataset comparisons of protein abundance, but also by providing the means to store and analyse proteomics data in contexts other than expression analyses. The protein-centric web interface provides researchers real-time and use-case-specific access to data for single or multiple proteins using interactive visualizations at different levels of detail. The data model of ProteomicsDB is able to store identification and quantification data from almost all conceivable proteomics experiments including meta-data such as sample preparation protocols, data acquisition parameters and sample treatment conditions. More recently, its capabilities have been expanded to also host results from other quantitative omics technologies ranging from drug-protein interaction studies and cell-viability experiments to data from public protein interaction databases and transcriptomes. In this article, we introduce the different analysis options available in ProteomicsDB and highlight the developments accumulated over the past three years.
RESULTS
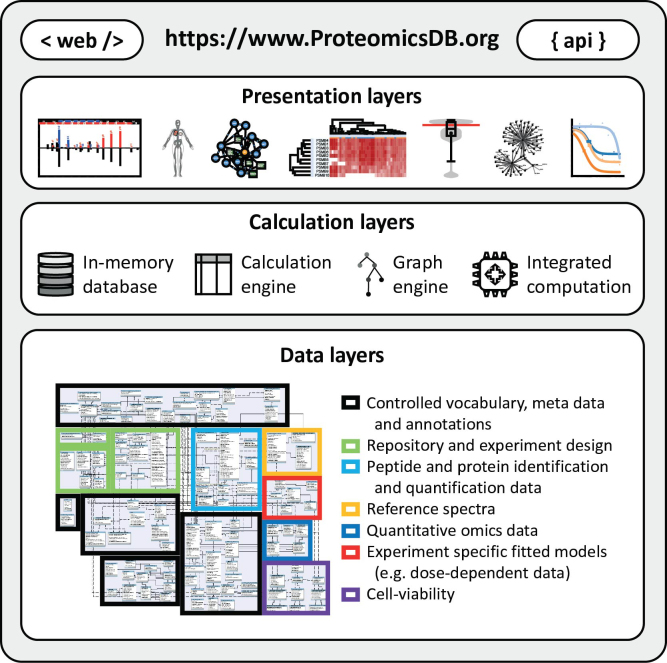
ProteomicsDB utilizes the in-memory database management system SAP HANA (11) and was developed to enable the real-time interactive exploration of large collections of quantitative mass spectrometry-based proteomics data (12). A major focus during the initial development of ProteomicsDB was to enable the storage of identification and quantification data on both peptide and protein level, irrespective of the experimental setup and analysis method used. Based on 408 experiments resulting from 78 experiments we identify 15721 of 19629 proteins covering 80% of the human proteome. A comparison to the Human Proteome Project (13,14) can be found in the Supplementary Table S1. To this end, ProteomicsDB is able to store the output of any algorithm used for the automatic interpretation of mass spectra (database search). Combined with the ability to map each observed peptide spectrum match (PSM) in any LC–MS/MS raw file transparently to the corresponding sample annotated with information on acquisition and sample preparation parameters, this ensures flexibility during data analysis. The storage of treatment conditions and the overall experimental design facilitate the analysis of more complex relations within and across different datasets, such as dose- and temperature-dependent assays. Efficient access to the data in combination with modern web-based visualization technologies facilitates real-time interactive exploration of heterogeneous data in an intuitive and simple way. All figures and tables available in ProteomicsDB can be downloaded, while an application programming interface allows users to directly interact with the database in order to download raw data for off-line processing or storage (Figure 1).
Figure 1.
ProteomicsDB consists of three major layers. The bottom layer is the data layer providing information to the calculation layer. It consists of seven major modules enabling the storage and retrieval of meta data, annotations and quantitative information associated with proteins and biological systems. Due to in-memory storage of the data layer, calculations using the calculation engine (structured query language), graph engine and other integrated programming languages (e.g. R and Python) are highly efficient. The results of these calculations can be explored in the presentation layer offering a variety of different interactive visualizations via the web interface or systematic access via the ProteomicsDB application programming interface (API).
Because of the in-memory capabilities of SAP HANA, most of the data shown on the website are not pre-computed, avoiding the need for monthly or yearly builds and enabling rapid adjustments. The different storage layers and versatile processing capabilities available in HANA enabled the integration of graph and standard relational database features. This facilitated the incorporation of many different data sources and led to the development of a variety of new features. While all protein-related results stored in ProteomicsDB are mapped to UniProt (15) identifiers, a versatile resource identifier mapping system enables a seamless conversion between different resources, which facilitates easy integration of additional data sources not mapped to UniProt (e.g. transcriptomics and interaction data).
In the following sections, we will start by briefly highlighting the data model used by ProteomicsDB and its developments over the past years. Subsequently, we will introduce the main features available on ProteomicsDB, which are organized in protein-centric visualizations for single and multiple proteins.
ProteomicsDB data model
The data model of ProteomicsDB is grouped into 7 major modules (Figure 1): (i) the meta-data, which contains annotations and ontologies; (ii) the repository, which contains the mapping of raw data to samples, experiments and projects, as well as associated meta-data and experimental designs; (iii) peptide and protein identification and quantification data, which stores spectra, the associated database search engine results, as well as peptide and protein abundance information; (iv) reference identification, which contains reference spectra from measurements of synthetic peptide standards; (v) the quantitative omics model; (vi) experiment specific models, such as dose-response models and (vii) cell-viability data. See Supplementary Text 1 for more details about the data models and its internal mechanisms.
Protein-centric web interface
ProteomicsDB is designed to enable researchers to quickly interrogate identification and quantification information of single and multiple proteins. For this purpose, ProteomicsDB offers two major ways to browse all available data. On the one hand, there is the presentation of information available for a single protein of interest. This can be accessed by either searching for a protein or peptide of interest in the ‘Human Proteins’ or ‘Peptides’ tab, respectively, or by browsing the human proteome in a ‘Chromosome’ centric view (Figure 2A). On the other hand, there are visualizations of specific aspects of the data for multiple proteins. This functionality is referred to as ‘Analytics’ within ProteomicsDB and can be found in the main menu at the top of the website. Currently, four analytical views are implemented and offer the cross-experiment analysis of protein expression, single and multiple drug selection and the exploration of cell viability data. It should be noted that ProteomicsDB is optimized for Firefox and Chrome.
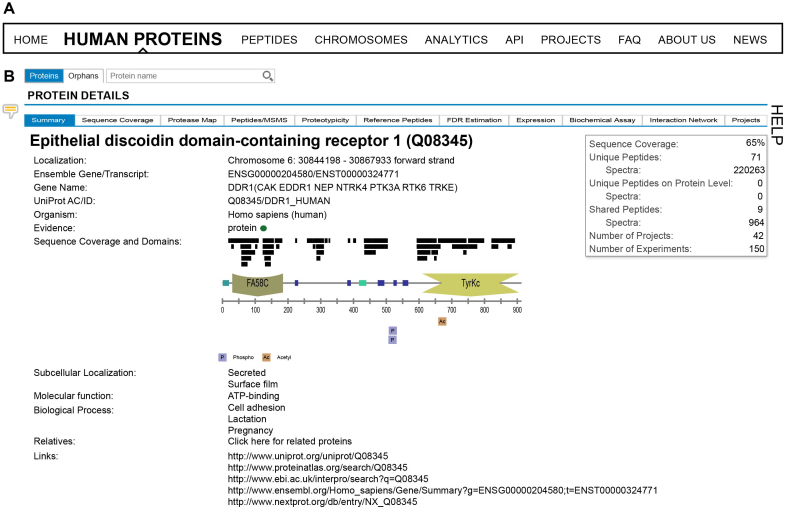
Figure 2.
(A) ProteomicsDB can be used to interrogate identification and quantification information on either single or multiple proteins. Information about single proteins can be accessed via the ‘Human Proteins’, ‘Peptides’, and ‘Chromosomes’ tabs. Information about multiple proteins can be explored via the ‘Analytics’ tab. (B) On the ‘Human Proteins’ tab, a brief summary is shown about the information available for a given protein. The corresponding domain structure is dynamically generated and alongside it, all observed peptides and post-translational modifications (PTMs) are displayed.
This section focuses on describing the visualizations for single (‘Human proteins’) and multiple proteins (‘Analytics’) in more detail. The features to analyse proteotypicity, reference peptides and FDR estimation for single proteins are fully described in Supplementary Text 2–4 (Supplementary Figures S1–4). Each page also provides a brief description of the functionalities by opening the ‘Help’-tab to the right of each page and tab. The feedback-icon, located on the left on each page, can be used to provide direct feedback, comments or report bugs to us. For the purpose of this paper, we will focus on one protein highlighting all available functions and visualizations throughout the manuscript. Discoidin Receptor 1 (DDR1) is a member of a family of receptor tyrosine kinases (RTKs) that is activated in response to collagen and is part of the arsenal of cell surface receptors that mediate tumor cell-environment interactions.
Human proteins
The search field can be used to browse proteins by gene name, accession number or protein description. The resulting table shows all available proteins partially matching the search string. All tables in ProteomicsDB can be filtered and sorted by clicking on a specific column header. Most tables also offer hiding or showing additional columns, which are not shown by default but are always included in downloaded csv files.
Protein summary
Upon selecting a protein of interest, the user sees a brief summary (Figure 2B) about the information available for the protein, including, but not limited to, the number of peptides which were detected (shared and unique on either gene or protein level), the sequence coverage and some basic annotations such as GO terms, chromosomal location, external links and evidence status. The evidence status is either red, yellow or green indicating missing, questionable and strong evidence for its identification, respectively. In addition, the domain structure of the protein is dynamically generated and shown in the middle of the page. Aligned to this, all observed peptides and post-translational modifications (PTMs) are visualized by black bars. This enables users to quickly investigate which part of the protein is (likely) ‘MS-accessible’ (i.e. produces peptides measureable by mass spectrometry) and which domains were previously observed to harbour post translational modifications (often an indicator of activity modulation). The sequence coverage view can be expanded to investigate which peptides were observed in detail. In addition, the ‘Sequence coverage’ tab can be opened to view the entire sequence of the protein. Stretches coloured in red indicate that this part is covered by peptides in ProteomicsDB. The theoretical sequence coverage can be explored using the ‘Protease map’ tab. One or several proteases can be chosen along with different peptide filter criteria, in order to predict which combination of proteases will lead to the highest (theoretical) cumulative sequence coverage. This feature can guide users in designing experiments that require high sequence coverage such as PTM or variant identification.
Peptides/MSMS
The ‘Peptides/MSMS’ tab can be used to check individual peptides and their corresponding spectra. The initial view lists all observed peptides including meta-data such as mass, length, uniqueness and the number of observations, as well as different measures of confidence, such as the search engine score. Each spectrum used for protein inference can be visualized in ProteomicsDB using the built-in spectrum browser. In order to view experimental spectra, an overlay containing all available PSMs for the selected peptide (Supplementary Figure S1 top table) can be opened by clicking a peptide of interest. Fragment ions in experimental spectra are annotated on request by an expert system (16). Annotation rules, such as calculated fragment ions and sequence-dependent neutral losses, are stored in the database and can be modified at any time. Annotation options for the spectrum, general visualization options and a fragmentation table can be opened to the left and right of the spectrum (Supplementary Figure S1).
An integrated feature of the spectrum viewer is the mirror representation of a reference spectrum (bottom spectrum) if available. These spectra originate from e.g. synthesized peptides, which were measured independently and can be used to validate the identification of peptides and in turn also proteins. This is especially useful when only a few peptides were identified for a specific protein, since such spurious identifications could originate from false matches during the database search. In case a reference spectrum for the selected peptide is available, the highest scoring PSM matching to the precursor charge and modification status of the selected PSM is chosen and displayed. Already today, ProteomicsDB stores more than 3 million reference spectra acquired as part of the ProteomeTools project (17) and covers more than 250k peptides measured in up to 11 different acquisition methods. For most peptides, multiple reference spectra are available and by default, the one acquired using similar acquisition parameters is shown. However, since parameters such as collision energy are not easily transferable between instruments (18), the user can choose to compare the experimental spectrum against any spectrum acquired under different conditions by selecting a different spectrum to the left of the spectrum viewer in the ‘Reference spectrum’ tab.
Expression
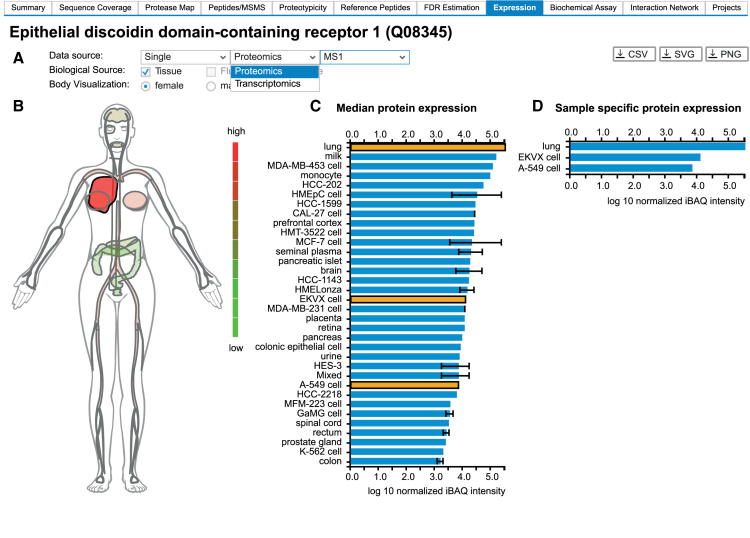
An essential feature of ProteomicsDB is the storage and visualization of quantitative data from a wide range of biological sources. While the initial development focused on the presentation of proteomics data, the generic implementation of ProteomicsDB also enables the storage and visualization of other omics data types, such as RNA-Seq data. The ‘Expression’ tab (Figure 3) can be used to explore the expression pattern of single proteins across the human body. The user can choose the primary data source and can visually explore the expression using a heatmap-like visualization of the human body. This view also superimposes abundance values of cell lines onto their respective tissue of origin and thus allows the integrated analysis of expression values originating from tissues or body fluids and cell lines.
Figure 3.
(A) ProteomicsDB can visualize expression data from different omics technologies. (B) A heatmap-like bodymap superimposing abundance values of tissues, fluids and cell lines (biological sources) onto their respective tissues of origin. (C) A bar chart resolving the expression data of b) on the level of their biological source. If multiple measurements for the same biological source are available, the error bar indicates the lowest and highest abundance observed for the selected protein. The bar chart and the bodymap are linked to each other, enabling the selection of either a tissue of origin in the bodymap (highlighted in dark red) or a biological source in the barchart (highlighted in orange). Here, the lung (high expression of DDR1), was selected in the bodymap, which automatically highlights all corresponding tissues and cell lines in the bar chart (EKVX cell and A-549 cell originated from lung tissue). (D) A bar chart visualizing sample-specific abundance values of the sources selected in middle bar chart (highlighted in orange). On click on one of the bars, the corresponding sample preparation protocol can be examined.
The expression view consists of two major components comprising data selection (Figure 3A) and visualization (Figure 3B–D). To enable meaningful cross-experiment comparison of expression values, only data from similar sources can be selected. For proteomics, MS1 and MS2 quantification techniques (19) cannot be compared directly, thus the filters only support the selection of either type. Likewise, the comparison of protein abundance measures originating from full proteome data (unbiased expression analysis) or affinity type experiments (biased abundance analysis) is not possible.
The data visualization is composed of three interactive and interconnected elements: (i) a heatmap-like body map (Figure 3B), (ii) a cell type aggregated bar chart (Figure 3C) and (iii) a sample specific bar chart (Figure 3D). The expression of DDR1 is restricted to epithelial cells, particularly in the kidney, lung, gastrointestinal tract and brain. Upon selection of a specific tissue in the heatmap, the middle barchart highlights all cell lines and tissues, which are connected to this tissue (e.g. tissue of origin). Likewise, the selection of a bar in the middle bar chart will highlight the corresponding tissue in the bodymap. This will also trigger the display of an additional bar chart, depicting the expression of the selected protein in a sample-specific manner. This view directly enables users to investigate the sample preparation and data acquisition parameters for each measurement by clicking on any bar in the bar chart on the right hand side.
Biochemical assay
Besides visualizing global expression patterns of proteins, ProteomicsDB is also able to make use of the stored experimental design to show changes in protein abundance upon specific treatments and sample handling steps. The ‘Biochemical assay’ tab (Supplementary Figure S3) provides dedicated views for such data and currently offers the exploration of Kinobeads (20,21) data, a competition binding assay used to decipher kinase:small molecule interactions, and two formats of cellular thermal shift data (22). Here, we will focus on the description of the Kinobeads data. Beyond this specific example, any relative protein abundance measured as a function of e.g. dose, temperature or time can be explored in the same way.
This view lists all available data for the selected protein, including direct and indirect targets as well as background proteins. In order to filter for binders, different filters are available and can be activated or deactivated. The slider can be used to filter for specific EC50 ranges or two goodness of fit values: R2 (R-square) and BIC (Bayesian information criterion). Dose response curves are fitted using a 4-parameter-log-logistic regression (23). Depending on the protein and the selected filters, the table will show multiple potential small molecules, which exhibit a dose-dependent effect. The experimental data is plotted using black circles, whereas the blue line shows the calculated dose response curve. The orange error bar spans ± one standard error of the EC50.
Interaction network
With the addition of protein–protein interaction (PPI) data from STRING and functional annotation data from KEGG, ProteomicsDB now offers interactive exploration of human PPI networks, enriched with functional information. This information can be accessed via the ‘Interaction Network’ tab (Supplementary Figure S4). We downloaded subscore-resolved protein network data, detailed interaction types as well as directionality information for Homo sapiens from STRING (24) and combined this data with pathway mappings obtained from KEGG (25) through their REST API. This information was mapped to canonical isoforms using UniProt. Therefore, selection of any protein isoform displays the PPI network and functional annotations of the corresponding canonical isoform. This protein-centric analysis allows the exploration of the PPI network with respect to a protein of interest (POI).
For each resource describing relations between proteins and/or functional categories incorporated into ProteomicsDB, it is possible to select only a subset of the included types of relations for visualization in the ‘Relations’ menu. This reduces the complexity of the PPI network and focuses the attention on relations of interest. Once all desired relation types for the different resources are selected from the menu on the left (not shown; available in the ‘Relations’ sub-tab), the interaction network for the selected protein can be displayed by clicking the ‘Start analysis’ button. This generates a force-directed graph in the network window to the right. Proteins and functional annotations are represented by circular and square nodes, respectively, while edges between nodes represent the relations between them. This initial graph only contains the POI together with the top five interacting proteins as determined by STRING subscore, all relations between them, as well as all functional annotations of the POI. Relations between two nodes without directionality information are merged into a single edge to further reduce redundancy in the graph. If desired, resources previously selected for visualization can be hidden by navigating to the ‘Options’ menu and toggling the corresponding radio buttons. At any point in time, the graph in the network window can be downloaded as a figure (.svg or .png) or a table (.sif) suitable for import into e.g. Cytoscape (26). An in-depth description to control the visualization can be found in Supplementary Text 5.
Analytics
So far, all analyses focused on the exploration of data relating to a single protein. The ‘Analytics’ section is designed to enable the analysis of data relating to multiple proteins. Currently, it offers four visualizations covering multi-protein expression pattern analysis, drug selection for single and combination treatments and the exploration of cell viability data.
Expression heatmap
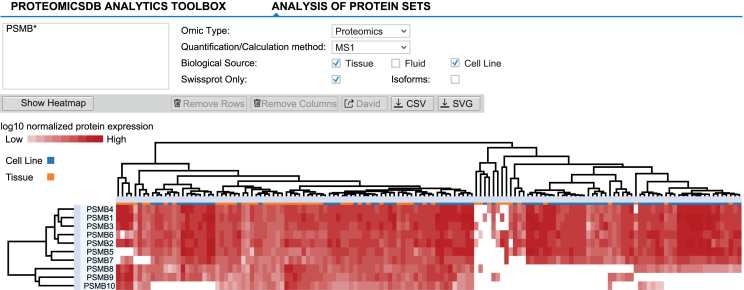
The comparison of protein expression profiles across different tissues, fluids and cell lines can give rise to new hypotheses and puts protein expression into context. While the expression tab of a single protein allows the analysis of expression patterns over multiple biological sources, it does not enable the analysis of multiple proteins simultaneously. This analysis is possible with the help of the ‘Expression heatmap’ tab (Figure 4), which shows proteins and biological sources as rows and columns, respectively. For this application, any list of gene names or UniProt identifiers can be supplied to ProteomicsDB. Similar to the ‘Expression’ tab, a user can choose between multiple available data sources and quantification methods.
Figure 4.
Expression heatmaps of multiple proteins across different tissues, fluids and cell lines can be displayed via the ‘Expression heatmap’ functionality of the ‘Analytics’ tab. Proteins and biological sources are shown as rows and columns, respectively. The dendrograms show the result of hierarchically clustering proteins and biological sources, respectively. Branches can be selected and either removed or used to perform GO-enrichment analyses (proteins). Here, all beta-units of the proteasome are displayed, suggesting differential expression of the canonical (expression of PSMB5, 6 and 7) and induced (expression of PSMB8, 9 and 10) proteasome across tissues and cell lines.
Figure 4 shows the resulting heatmap when searching for beta subunits of the proteasome ‘PSMB*’ in tissues and cell lines using protein expression values estimated by the iBAQ approach. The heatmap is fully interactive and provides multiple options to adjust and explore the data. Additional features of the heatmap are explained in Supplementary Text 6.
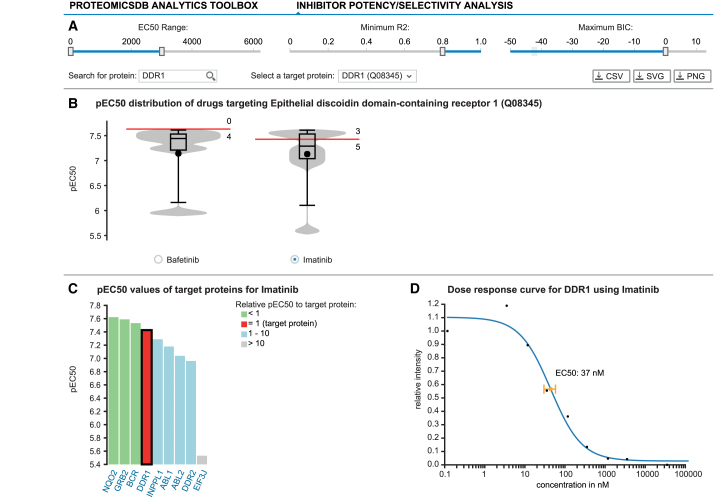
Inhibitor potency/selectivity analysis
One topic of great scientific interest is finding the most selective and potent drug against a specific target of interest. For this purpose, ProteomicsDB enables the interactive exploration of dose-dependent competition-binding data in a multi-protein-centric view (Figure 5). Starting with the selection of a protein of interest (here DDR1) the user can filter models based on dose-dependent data available for this protein using several criteria: the EC50 range, the R2 and BIC (similar as in the ‘Biochemical assay’ tab) (Figure 5A). The pEC50 (–log10 EC50 in nM) distribution of all targets meeting the filter criteria for each drug showing a dose-dependent effect on the selected target are plotted in separate violin charts (Figure 5B). The red marker indicates the EC50 of the selected protein for each drug. The selectivity of each compound can be evaluated by the numbers above and below the red marker, which depict the number of targets with higher or lower potency compared to the selected protein, respectively.
Figure 5.
ProteomicsDB enables the exploration of drug selectivity data from various sources. (A) Starting with the selection of a target protein, the user can filter fitted selectivity curves using several criteria: the EC50 range, the R2 and BIC. (B) Violin plots depicting the pEC50 (-log10 EC50) distributions for all compounds targeting the selected protein given the filter criteria from (A). The red marker indicates the EC50 of the selected protein for each drug. Numbers above and below the red marker indicate the number of other target proteins with higher or lower potency, respectively. At the time of writing, Bafetinib shows the most potent and selective inhibition of DDR1 with the given filters. (C) Bar chart displaying the distribution of pEC50 values for Imatinib depicting all of its protein:drug interactions available in ProteomicsDB. (D) The underlying raw data and the fitted model can be investigated on click on one of the bars (black border). The scatter plot highlights the EC50 for the selected protein:drug pair.
Users can inspect the pEC50 distribution of all targets for a given drug in an ordered bar chart by selecting the radio button underneath the corresponding violin plot. Targets depicted in (i) green, (ii) blue and (ii) gray are (a) more potent, (b) have similar potency or are (c) at least 10× less potent than the selected target (red), respectively (Figure 5C). This bar chart enables the investigation of all other targets of the selected drug, which could—depending on its use—increase the risk of unwanted side effects. Individual dose–response plots can be investigated by selecting a specific drug:protein interaction in the bar chart. On click—similar to the ‘Biochemical assay’ tab—a scatter plot depicting the individual measurements (black dots) and the fitted dose-response model (blue curve) with its estimated EC50 and standard error is shown to the right of the bar chart (Figure 5D).
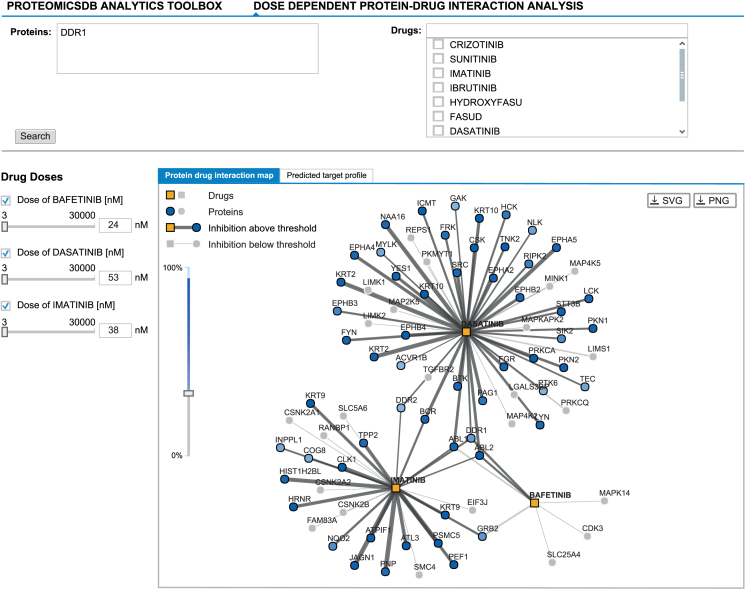
Dose-dependent protein-drug interaction analysis
The potency analysis provides an interface to select an inhibitor for a single protein of interest. However, in some applications, targeting multiple proteins can lead to a more effective treatment (e.g. to suppress resistance formation). The ‘Dose-dependent protein–drug interaction analysis’ tab (Figure 6) provides an interface to explore the predicted dose-dependent effects of multiple drugs on multiple proteins. This enables the selection of the most promising drug-combination to inhibit a set of proteins, while maintaining the lowest number of off-targets to decrease the chances of unwanted side-effects. Two views are available, which show the predicted target profile of the selected drugs at a certain dose as (i) a protein–drug interaction graph and (ii) a table showing the predicted inhibition effects. Both views are based on the dose-dependent models stored in ProteomicsDB.
Figure 6.
The ‘Dose-dependent protein-drug interaction analysis’ enables exploring protein:drug interaction data in a multi-drug fashion. It allows the selection of promising drug combinations suitable to inhibit a given target protein (here DDR1). The graph-view shows the protein-drug interaction landscape of selected drugs. Drugs (squares) and proteins (circles) are connected if binding/inhibition curves (‘Biochemical Assay’ data) are available. Predicted inhibitory effects are highlighted in the graph by dark grey edges of varying thickness (proportional to the EC50) and proteins coloured in different shades of blue (indicates the level of inhibition). Predicted inhibitory effects are only shown in case they surpass a user-defined cutoff (left vertical slider). The concentration of a drug can be adjusted by either clicking an edge (sets the concentration of the drug to the EC50 of that interaction), by manually adjusting the concentration using the sliders on the left or by entering the desired concentration into the textbox (left; next to sliders). Again, Bafetinib shows the most selective inhibition of DDR1 at an EC50 of 24 nM in comparison to the other two available inhibitors Imatinib (38 nM) and Dasatinib (53 nM).
The “Proteins" search field accepts sets of protein names. On this basis, all drugs showing at least one inhibitory effect on one of the proteins are taken into consideration. Alternatively, the “Drugs" search field can be used to manually add/select a set of drugs. In case both fields are used, the union of all drugs, either inhibiting at least one of the target proteins or selected manually, is used.
The graph-view shows the protein-drug interaction landscape of the selected drugs. Proteins (circles) are connected to drugs (squares) if a binding/inhibition curve is available for this combination. Each drug selected for the analysis is displayed on the left hand side of the view. The checkbox can be used to disable (hide) a drug from both views. In addition, the dose of each drug can be adjusted by moving the slider or by manually entering a desired drug-concentration. The predicted inhibition of a particular protein in both the graph and the table view are updated in real-time based on the given concentration of a drug. Predicted inhibitory effects are highlighted in the graph by grey edges of varying thickness (proportional to EC50) and blue proteins, the shading of which indicates the level of inhibition. Predicted inhibitory effects are only shown in case they surpass a user-defined cutoff (left vertical slider). In addition to the manual drug concentration, selecting an edge between a protein:drug pair sets the concentration of the drug to the EC50 of that interaction.
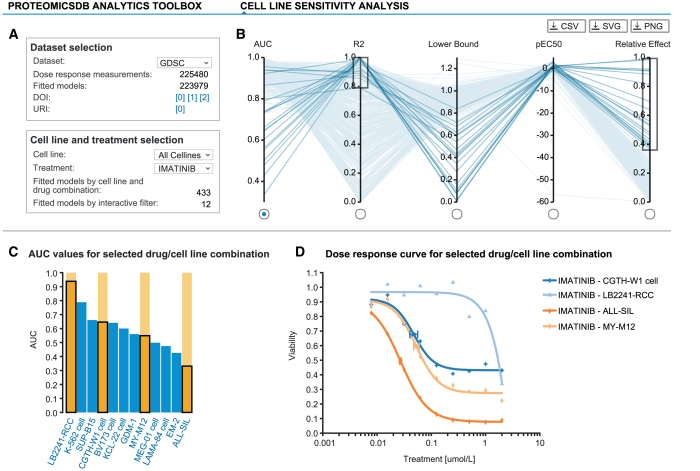
Cell viability data exploration
With the inclusion of dose-resolved viability data from several large-scale drug sensitivity studies (27–30), ProteomicsDB is now providing tools for fast exploration of dose-response curves quantifying sensitivity and resistance of hundreds of cell lines across hundreds of inhibitors (Figure 7). For each dose-response dataset, ProteomicsDB offers inhibitor- and cell line-centric analysis tools, which allow the identification of sensitive/resistant cell lines for a given inhibitor, while also enabling the identification of potent/impotent inhibitors for a given cell line, respectively. We downloaded dose-resolved viability data from various sources and converted them to relative viabilities, in order to bring the different datasets onto the same scale. Subsequently, the classical symmetric four-parameter log-logistic model was fitted to each inhibitor/cell line combination in each dataset, followed by parameter extraction and calculation of several summary statistics.
Figure 7.
ProteomicsDB incorporates several publicly available large-scale drug sensitivity screens. (A) Each drug sensitivity dataset in ProteomicsDB can be explored in a cell-line- or inhibitor-centric way and general statistics are shown for a given selection. (B) Users can interactively filter dose-response models based on multiple parameters such as AUC, R2, lower bound, pEC50 and relative effect (percent decrease in viability over the tested concentration range). (C) The distribution of a given parameter is visualized in a bar chart on selection of an axis in (B). (D) The underlying raw and fitted data can be investigated on click on one or many of the bars (highlighted in orange). The scatter plot highlights the EC50 for the selected cell line:drug pairs. The cell lines CGTH-W1, LB2241-RCC, ALL-SIL and MY-M12 show a clear dose-dependent effect on their viability upon Imatinib treatment. However, their EC50 values vary, highlighting that these cell lines show differential sensitivity/resistance to Imatinib.
After selecting a dataset of interest, analyses can be either cell line- or inhibitor-centric. For this purpose, either one cell line can be chosen, comparing all available inhibitors on this single cell line, or, vice versa, an inhibitor can be selected in order to compare the viabilities of all tested cell lines (Figure 7A). Selection of one cell line and one inhibitor is also possible, enabling direct investigation of a specific cell line/inhibitor pair. Using a parallel coordinates plot, it is possible to filter for multiple model parameters and different summary statistics simultaneously (Figure 7B). The exact distribution for each of these variables can be investigated across all cell lines/inhibitors, while selection of one or more cell lines/inhibitors allows the inspection of the underlying dose-resolved viability data (Figure 7C). Visualization of dose-resolved data for multiple cell lines/inhibitors—while essential for judging the reliability of the experimental data—is a feature largely missing from the web portals associated with the original publications (Figure 7D).
FUTURE DIRECTIONS
The large collection of experimental and reference spectra stored in ProteomicsDB opens the door for the development of new functionalities. For example, since the ProteomeTools project covers the entire human proteome with reference spectra, a systematic orthogonal evaluation of protein FDR using synthetic spectra becomes possible. This will provide further validation of protein identification events in ProteomicsDB. Similarly, spectra in ProteomicsDB could be downloaded or compared directly to user data in order to validate the identification of proteins with no prior observations. Furthermore, the combination of reference and experimental spectra and their chromatographic properties will enable the development of tools to guide the development of directed and targeted experiments by custom data-driven spectral library generation. Such tools could make use of the cell line- and tissue-specific protein background identified before and could provide experiment-driven estimates of interfering peptides.
Due to ProteomicsDB’s in-memory architecture, performing database-wide protein inference on all proteins at the same time is possible. This was essential in the development of the picked-FDR approach (31). While a significant proportion of the data stored in ProteomicsDB is already programmatically accessible, an obvious next step is the extension of this service to enable systematic access to all data. By extending the accessibility of data, ProteomicsDB might become an important infrastructure for computational scientists to develop and test new algorithms and for biologists to generate and test new hypotheses.
We will further broaden the scope of ProteomicsDB over the next years. One of the upcoming extensions is to provide protein abundance estimates for other organisms, such as Mus musculus and Arabidopsis thaliana. While the data model already supports the import of data from other model organisms, the user interface will need to be adjusted. With the ability to store expression patterns from other organisms, cross-species comparisons becomes possible enabling a plethora of questions to be asked and answered.
With the integration of other data sources into ProteomicsDB, the comparative visualization of multiple orthogonal dataset is within reach. We have already started to cross-link visualization tools within ProteomicsDB in the ‘Interaction network’ tab. However, the integrated data model of ProteomicsDB will also enable the interactive visualization of multiple data sources at once in order to provide a more comprehensive view on omics data. For example, the annotation rows in the ‘Expression heatmap’ can be extended to show drug selectivity data for proteins or memberships in pathways and signalling cascades, while the annotation columns can show cell viability data for biological sources. Similarly, the ‘Expression heatmap’ could make use of the imported protein–protein interaction and pathway data and allow users to add proteins to the heatmap based on the network neighborhood of the selected protein. Integrated models of drug-selectivity, cell-viability and protein/mRNA expression could be trained to predict treatment outcomes and estimate missing values in either dataset. Given the computational power of the underlying hardware, it is even conceivable to provide the infrastructure and interfaces to users to upload their own data for direct comparison and for direct model training on their phenotypic measurements (32).
AVAILABILITY
ProteomicsDB is available under https://www.ProteomicsDB.org.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank all previous members of the ProteomicsDB project team, Hannes Hahne (TUM), Adelya Fatykhova, Anja Gerstmair, David Weese, Emanuel Ziegler, Franz Faerber, Helmut Cossmann, Ingrid Hurbain, Jan Huenges, Joos-Hendrik Boese, Lars Butzmann, Lars Rueckert, Marcus Lieberenz, Mohammed AbuJarour, Wilhelm Becker (SAP), Marcus Bantscheff, Mikhail Savitski, Toby Mathieson (GSK), and the Kuster laboratory for fruitful discussions and technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
German Federal Ministry of Education and Research (BMBF) [031L0008A] (in part). Funding for open access charge: German Federal Ministry of Education and Research (BMBF) [031L0008A].
Conflict of interest statement. M.W. and B.K. are founders and shareholders of OmicScouts, which operates in the field of proteomics. They have no operational role in the company and their involvement had no impact on the current study. S.G., J.S., H.-C.E. and S.A. are employees of SAP SE.
REFERENCES
- 1. Aebersold R., Mann M.. Mass spectrometry-based proteomics. Nature. 2003; 422:198–207. [DOI] [PubMed] [Google Scholar]
- 2. Han X., Aslanian A., Yates J.R. 3rd. Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008; 12:483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hawkridge A.M., Muddiman D.C.. Mass spectrometry-based biomarker discovery: toward a global proteome index of individuality. Annu. Rev. Anal. Chem. (Palo Alto Calif.). 2009; 2:265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Riffle M., Eng J.K.. Proteomics data repositories. Proteomics. 2009; 9:4653–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perez-Riverol Y., Alpi E., Wang R., Hermjakob H., Vizcaino J.A.. Making proteomics data accessible and reusable: current state of proteomics databases and repositories. Proteomics. 2015; 15:930–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vizcaino J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Desiere F., Deutsch E.W., King N.L., Nesvizhskii A.I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S.N., Aebersold R.. The PeptideAtlas project. Nucleic Acids Res. 2006; 34:D655–D658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Craig R., Cortens J.P., Beavis R.C.. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 2004; 3:1234–1242. [DOI] [PubMed] [Google Scholar]
- 9. Wang M., Weiss M., Simonovic M., Haertinger G., Schrimpf S.P., Hengartner M.O., von Mering C.. PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell Proteomics. 2012; 11:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schaab C., Geiger T., Stoehr G., Cox J., Mann M.. Analysis of high accuracy, quantitative proteomics data in the MaxQB database. Mol. Cell Proteomics. 2012; 11, doi:10.1074/mcp.M111.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Färber F., Cha S.K., Primsch J., Bornhövd C., Sigg S., Lehner W.. SAP HANA database. ACM SIGMOD Record. 2012; 40:45–51. [Google Scholar]
- 12. Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H. et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014; 509:582–587. [DOI] [PubMed] [Google Scholar]
- 13. Legrain P., Aebersold R., Archakov A., Bairoch A., Bala K., Beretta L., Bergeron J., Borchers C.H., Corthals G.L., Costello C.E. et al. The human proteome project: current state and future direction. Mol. Cell Proteomics. 2011; 10, doi:10.1074/mcp.M111.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaudet P., Michel P.A., Zahn-Zabal M., Britan A., Cusin I., Domagalski M., Duek P.D., Gateau A., Gleizes A., Hinard V. et al. The neXtProt knowledgebase on human proteins: 2017 update. Nucleic Acids Res. 2017; 45:D177–D182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neuhauser N., Michalski A., Cox J., Mann M.. Expert system for computer-assisted annotation of MS/MS spectra. Mol. Cell Proteomics. 2012; 11:1500–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zolg D.P., Wilhelm M., Schnatbaum K., Zerweck J., Knaute T., Delanghe B., Bailey D.J., Gessulat S., Ehrlich H.C., Weininger M. et al. Building ProteomeTools based on a complete synthetic human proteome. Nat. Methods. 2017; 14:259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zolg D.P., Wilhelm M., Yu P., Knaute T., Zerweck J., Wenschuh H., Reimer U., Schnatbaum K., Kuster B.. PROCAL: A set of 40 peptide standards for retention time indexing, column performance monitoring and collision energy calibration. Proteomics. 2017; doi:10.1002/pmic.201700263. [DOI] [PubMed] [Google Scholar]
- 19. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B.. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007; 389:1017–1031. [DOI] [PubMed] [Google Scholar]
- 20. Lemeer S., Zorgiebel C., Ruprecht B., Kohl K., Kuster B.. Comparing immobilized kinase inhibitors and covalent ATP probes for proteomic profiling of kinase expression and drug selectivity. J. Proteome Res. 2013; 12:1723–1731. [DOI] [PubMed] [Google Scholar]
- 21. Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C. et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 2007; 25:1035–1044. [DOI] [PubMed] [Google Scholar]
- 22. Savitski M.M., Reinhard F.B., Franken H., Werner T., Savitski M.F., Eberhard D., Martinez Molina D., Jafari R., Dovega R.B., Klaeger S. et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science. 2014; 346:1255784. [DOI] [PubMed] [Google Scholar]
- 23. Ritz C., Streibig J.C.. Bioassay Analysis using R. J. Stat. Softw. 2005; 12, doi:10.18637/jss.v012.i05. [Google Scholar]
- 24. Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M. et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009; 37:D412–D416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H. et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016; 166:740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B. et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat. Commun. 2015; 6:7002. [DOI] [PubMed] [Google Scholar]
- 30. Rees M.G., Seashore-Ludlow B., Cheah J.H., Adams D.J., Price E.V., Gill S., Javaid S., Coletti M.E., Jones V.L., Bodycombe N.E. et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016; 12:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Savitski M.M., Wilhelm M., Hahne H., Kuster B., Bantscheff M.. A Scalable Approach for Protein False Discovery Rate Estimation in Large Proteomic Data Sets. Mol. Cell Proteomics. 2015; 14:2394–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gujral T.S., Peshkin L., Kirschner M.W.. Exploiting polypharmacology for drug target deconvolution. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5048–5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.