Abstract
Transcription factors (TFs) are major trans-acting factors in transcriptional regulation. Therefore, elucidating TF–target interactions is a key step toward understanding the regulatory circuitry underlying complex traits such as human diseases. We previously published a reference TF–target interaction database for humans—TRRUST (Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining)—which was constructed using sentence-based text mining, followed by manual curation. Here, we present TRRUST v2 (www.grnpedia.org/trrust) with a significant improvement from the previous version, including a significantly increased size of the database consisting of 8444 regulatory interactions for 800 TFs in humans. More importantly, TRRUST v2 also contains a database for TF–target interactions in mice, including 6552 TF–target interactions for 828 mouse TFs. TRRUST v2 is also substantially more comprehensive and less biased than other TF–target interaction databases. We also improved the web interface, which now enables prioritization of key TFs for a physiological condition depicted by a set of user-input transcriptional responsive genes. With the significant expansion in the database size and inclusion of the new web tool for TF prioritization, we believe that TRRUST v2 will be a versatile database for the study of the transcriptional regulation involved in human diseases.
INTRODUCTION
Transcriptional regulation is a key process that determines the developmental fate and cellular responses to genetic and environmental perturbation. Some of the most important trans-acting factors involved in this process are transcription factors (TF), which bind to cis-regulatory elements of the DNA and activate RNA polymerase to begin the transcription of target genes. Activation of specific TFs can induce differentiation into certain cell types, and mutations in TFs cause dysregulation of target genes as well as their downstream genes, often resulting in disease states. Researchers have studied interactions between TFs and their target genes for many years to reconstruct transcriptional regulatory circuitries in cell types under various spatiotemporal contexts by using experimental and computational approaches. However, the complexity of regulatory circuitries is overwhelming, and most, if not all, technologies currently available are limited in accuracy as well as comprehensiveness. To evaluate reconstructed transcriptional regulatory networks, we first need to establish a highly accurate and comprehensive database of reference TF–target regulatory interactions.
Therefore, we previously constructed and published TRRUST (1), a database of reference TF–target regulatory interactions in humans based on literature curation. We conducted sentence-based text mining and prioritized the candidate sentences for the cost-effective literature curation. From manual curation of 23 409 candidate sentences, we were able to retrieve 8015 TF–target interactions for 748 human TFs. Significant efforts have been made in the past to retrieve information regarding the mode of regulation (MoR) for the interactions: either activation or repression. In our previous study, we could provide MoR information for ∼60.6% of the interactions in TRRUST. Furthermore, TRRUST was the most comprehensive public database for literature-curated TF–target interactions in humans. We also showed that TRRUST could benchmark transcriptional regulatory networks inferred from high-throughput genomics data.
In the present study, we present TRRUST v2 (version 2), which is a significantly improved version in terms of information content and its utility. TRRUST v2 (www.grnpedia.org/trrust) contains 8444 human TF–target interactions for 800 human TFs, which is significantly higher than the amount of information available in the previous version. Most importantly, we have also added 6552 TF–target interactions for 828 mouse TFs to the database. Mice are the most important and commonly used laboratory animal for biomedical research. Moreover, comparison of regulatory networks between humans and mice has revealed a high degree of conservation of trans-regulatory features including regulatory network architecture (2). Therefore, we expect that the information regarding mouse TF–target interactions will effectively complement the information on human regulatory interactions. We also compared TRRUST v2 to other literature-curated databases containing TF–target interactions and found it to be superior to all others for both human and mouse information. Finally, to improve the utility of the TF–target interactions for elucidating transcriptional regulatory circuits involved in diseases, we implemented a network-based algorithm to prioritize key TFs for the given transcriptionally responsive genes. Using this prediction tool, we could correctly retrieve a perturbed TF and successfully predict candidate TFs for lung cancers that could be validated by literature. These results indicate that TRRUST v2 is a versatile database to study the transcriptional regulation involved in human diseases.
DATABASE IMPROVEMENT
Overview of the literature curation process for mouse TF–target interactions
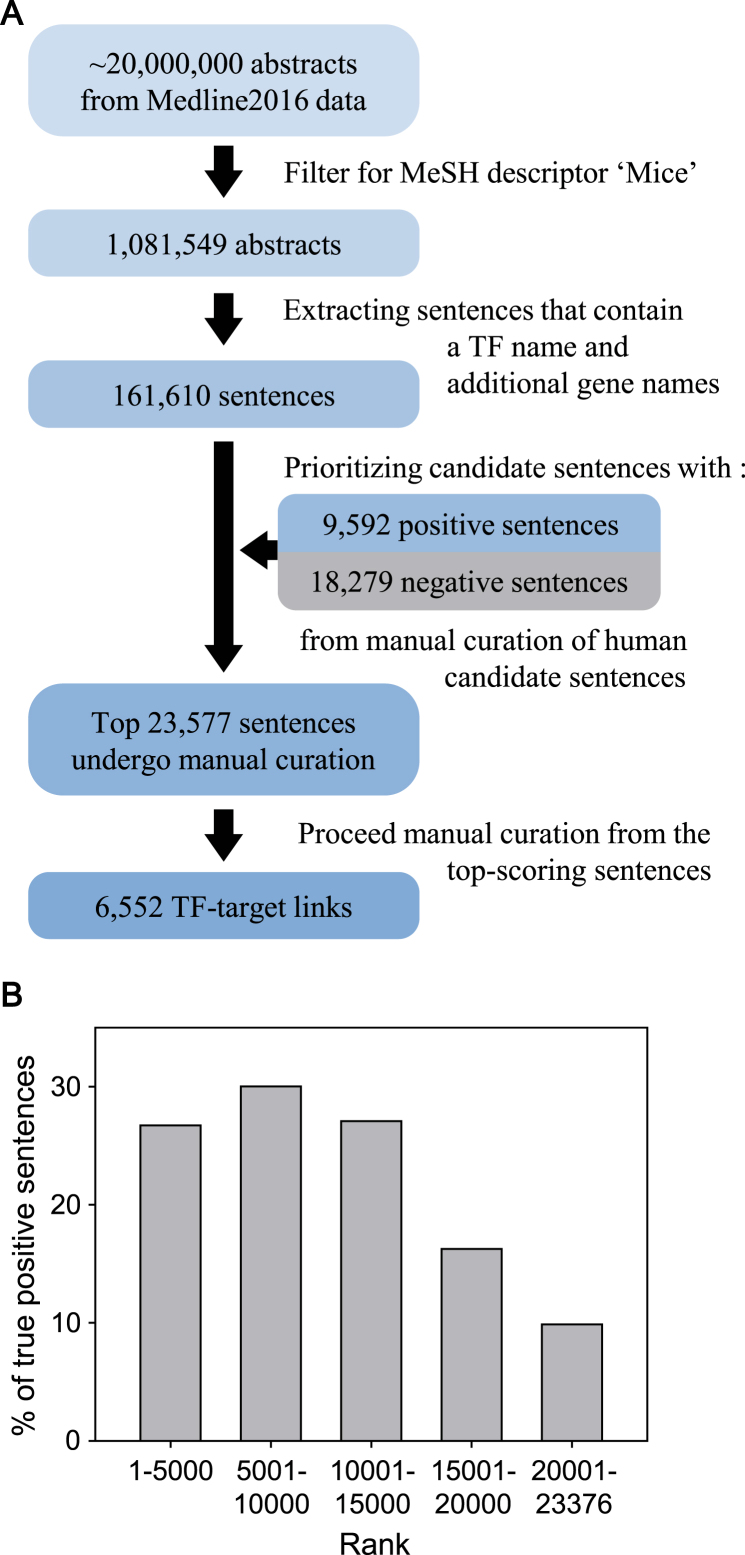
The overall process of retrieving mouse TF–target interactions from the literature is summarized in Figure 1A. The Medline database holds more than 20 million abstracts from life sciences and biomedical articles. We first filtered 1 081 549 abstracts from Medline2016 data using the MeSH descriptor ‘Mice’. In the filtered abstracts, we scanned for sentences that contained at least one TF name and additional gene names. We used a list of 1743 mouse TF genes compiled by Ravasi et al. (3) and a list of mouse gene names and their synonyms derived from the NCBI Gene database (4), which were further filtered for the Consensus coding sequences (CCDS) database (5). As a result of these filtration process, we collected a total of 161 610 candidate sentences for the following step of the literature curation process.
Figure 1.
(A) An overall workflow of sentence-based text mining for mouse TF–target regulatory interactions from research articles. (B) Proportion of true-positive sentences for each bin of 5000 candidate sentences rank-ordered by scores based on the difference between the frequencies of each word based on positive gold-standard sentences and that based on negative gold-standard sentences.
It was possible that the filtered sentences contained appropriate gene names, but did not have an indication of the regulatory interactions. Moreover, exhaustive manual curation for all the 161 610 sentences would be a daunting task. We therefore prioritized the sentences using methods similar to those described in our previous work (1). Using all the sentences subjected to manual curation for human TF–target interactions, we then established a set of 9592 positive gold-standard sentences (i.e. information of TF–target interactions was confirmed) and a set of 18 279 negative gold-standard sentences (i.e. manual curation revealed no information of TF–target interaction). We then prioritized the candidate sentences for transcriptional regulation in mice based on the sum of the difference of the frequency of occurrence of each word between the positive and negative gold-standard sentences. The score S for a candidate sentence composed of n words was calculated using the following equation:
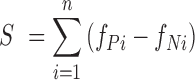 |
where 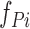 and
and 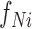 are the frequencies of the ith word of the candidate sentence among the positive and negative gold-standard sentences, respectively. If the words of a candidate sentence tended to have higher frequencies among the positive gold-standard sentences than among the negatives, the candidate sentence was more likely to have information regarding TF–target interactions.
are the frequencies of the ith word of the candidate sentence among the positive and negative gold-standard sentences, respectively. If the words of a candidate sentence tended to have higher frequencies among the positive gold-standard sentences than among the negatives, the candidate sentence was more likely to have information regarding TF–target interactions.
We then proceeded with manual curation from the top-scored candidate sentences. During manual curation, we examined not only candidate sentences but also the abstracts that included the sentences to confirm the retrieved information in the larger textual context. Sentences for molecular interactions involved in processes other than transcriptional regulation, such as protein–protein interactions and interactions with chemicals, were excluded. Sentences were also excluded if either gene name for the interaction was not annotated by CCDS annotations (5). At least one experienced curator confirmed the retrieved TF–target interactions from the sentences. A total of 23 577 sentences were subjected to manual curation for constructing the mouse database in TRRUST v2. We found some discrepancy between MeSH information and the abstract content regarding organisms. For example, some abstracts clearly indicated the given TF–target interactions occurred in mice but were tagged by MeSH terms of both ‘humans’ and ‘mice’. Therefore, we examined full-text articles if a given TF had the same name but with a different letter case between two species to confirm organism assignment, consequently requiring full-text inspection for more than 1200 articles.
Next, to evaluate the effectiveness of the scoring scheme for prioritizing the candidate sentences, we examined the proportion of true positive sentences that harbored information regarding mouse TF–target interactions for each bin of 5000 sentences from the highest score. We observed a significantly higher proportion of true positive sentences from the top three bins than from others with lower scores (Figure 1B). We could retrieve mouse TF–target interactions from 27–30% of the 5000 sentences from the top three bins. However, the proportion of true positive sentences dropped to 16 and 10% for the subsequent bins with lower scores. Given the significant decrease in the proportion of true positive sentences, we froze the current version of mouse TF–target interaction database based on the top 23 577 sentences, which yielded 6552 TF–target interactions for 828 mouse TFs. These results indicated that our strategy of prioritizing candidate sentences was useful for cost-effective manual curation as it effectively differentiated sentences for their likelihood of containing information regarding TF–target interactions. Manual curation of the remaining candidate sentences will be continuously conducted to expand the database in the future.
Expansion of the database for human TF–target interactions
In addition to the new mouse TF–target interactions, we also expanded the size of the database for human TF–target interactions in TRRUST v2. We previously generated positive and negative gold-standard sentences for humans via initial manual curation of 17 409 out of 57 360 candidate sentences. Using the scoring scheme described above, we then prioritized the remaining candidate sentences and continued manual curation for the top 6000 candidate sentences (i.e. a total of 23 409 candidate sentences were curated); this yielded 8015 human TF–target interactions for the previous version of TRRUST. However, since the publication of the first version of TRRUST, we have performed manual curation for an additional top 4,000 candidate sentences for humans. As a result, TRRUST v2 now contains a total of 8444 TF–target interactions for 800 human TFs, which is approximately a 5.3 and 6.9% increase in the number of interactions and TFs, respectively.
DATABASE ASSESSMENT
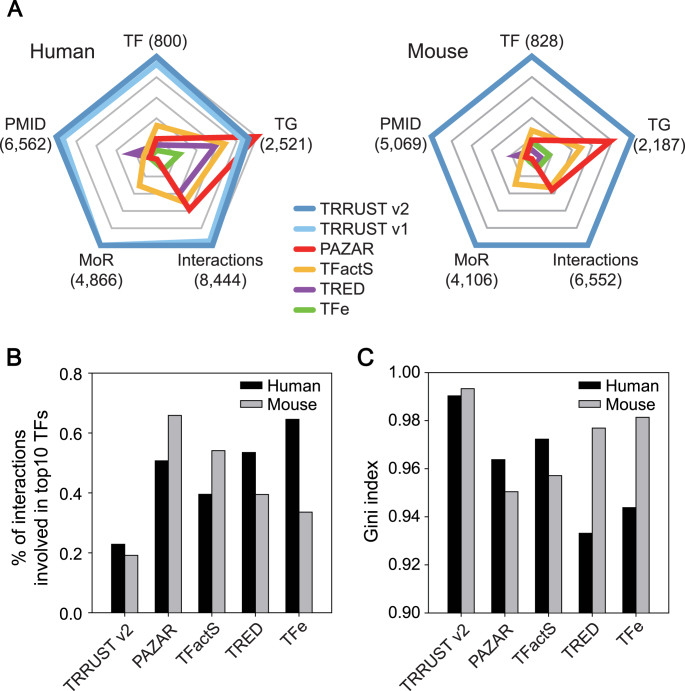
Based on the combined count of 14 996 human and mouse TF–target interactions, TRRUST v2 contains more than twice as much information as the previous version. We also compared the improved TRRUST v2 with other manually curated databases for TF–target interactions. We compiled information regarding regulatory interactions from PAZAR (6), TFactS (7), TRED (8) and TFe (9) and then compared it with TRRUST v2 in terms of the number of TFs, targets, interactions, interactions with MoR and supporting PubMed articles. Besides TRRUST v2, only TFactS and TFe provided MoR information. For TRED, we considered interactions with a binding quality of ‘known’ as literature-curated interactions. We found that TRRUST v2 to be the most comprehensive database for both human and mouse regulatory interactions (Table 1 and Figure 2A). Moreover, the number of human TF–target interactions in TRRUST v2 is twice that of the next biggest database, PAZAR.
Table 1. Summary of databases of transcription factor (TF)–target interactions.
| Human | Mouse | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TF | TG | Interaction | MoR | PMID | TF | TG | Interaction | MoR | PMID | |
| TRRUST v2 | 800 | 2521 | 8444 | 4866 | 6562 | 828 | 2187 | 6552 | 4106 | 5069 |
| TRRUST v1 | 748 | 2375 | 8015 | 4861 | 6175 | |||||
| PAZAR | 173 | 2747 | 4918 | 489 | 162 | 1726 | 2329 | 328 | ||
| TFactS | 275 | 1873 | 4209 | 1468 | 776 | 238 | 1080 | 2149 | 1176 | 472 |
| TRED | 119 | 1621 | 3275 | 1542 | 75 | 236 | 508 | 745 | ||
| TFe | 76 | 623 | 1058 | 134 | 510 | 147 | 388 | 673 | 90 | 353 |
TF, the number of TF; TG, the number of target genes; Interaction, the number of TF–target interactions; MoR, the number of TF–target interactions with mode of regulation information; PMID, the number of PubMed articles to support the given database.
Figure 2.
Comparison of TRRUST v2 with other databases of TF–target interactions. (A) Radar plots depicting five database-related parameters: TF, the number of transcription factors (TFs); TG, the number of target genes; Interaction, the number of TF–target interactions; MoR, the number of TF–target interactions with mode of regulation information; PMID, the number of PubMed articles to support the given database. The maximum value of each parameter is shown in parentheses, and scales were normalized by the maximum value. (B) The proportion of the interaction involving the top 10 TFs with the highest number of interactions in humans and mice. (C) Gini index that measures the degree of dispersion of interactions across TFs in humans and mice.
The amount of information may not be a sufficient criterion to determine a database as useful. If the majority of TF–target interactions of the database are concentrated in only a minor group of TFs, the usefulness of the database could be unsatisfactory. Given that there exist study and technical biases in biomedical research, the information extracted from research articles can be easily biased toward some genes with higher clinical importance or toward those with well-developed experimental methods for investigation. Therefore, we first examined the proportion of total regulatory interactions involving the top 10 TFs for each database (Figure 2B) and found that the top 10 TFs of TRRUST v2 are involved in 22 and 19% of the total human and mouse regulatory interactions, respectively. However, the top 10 TFs of other public databases turned out to be involved in a substantially higher proportion of the total regulatory interactions. We also examined the degree of dispersion of the regulatory interactions across TFs in each database using the Gini index (G) as follows:
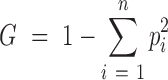 |
where 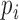 is the proportion of interactions involving the
is the proportion of interactions involving the 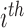 TF in the ranks from the largest number of interactions, and n is the number of TFs involved in the interactions. TRRUST v2 showed the highest Gini index, which indicates the most dispersed regulatory interactions across TFs (Figure 2C). These results indicated that the database of transcriptional regulatory interactions in TRRUST v2 is not only more comprehensive but also less biased than other public databases.
TF in the ranks from the largest number of interactions, and n is the number of TFs involved in the interactions. TRRUST v2 showed the highest Gini index, which indicates the most dispersed regulatory interactions across TFs (Figure 2C). These results indicated that the database of transcriptional regulatory interactions in TRRUST v2 is not only more comprehensive but also less biased than other public databases.
WEB INTERFACE IMPROVEMENT
Web interface for mouse database
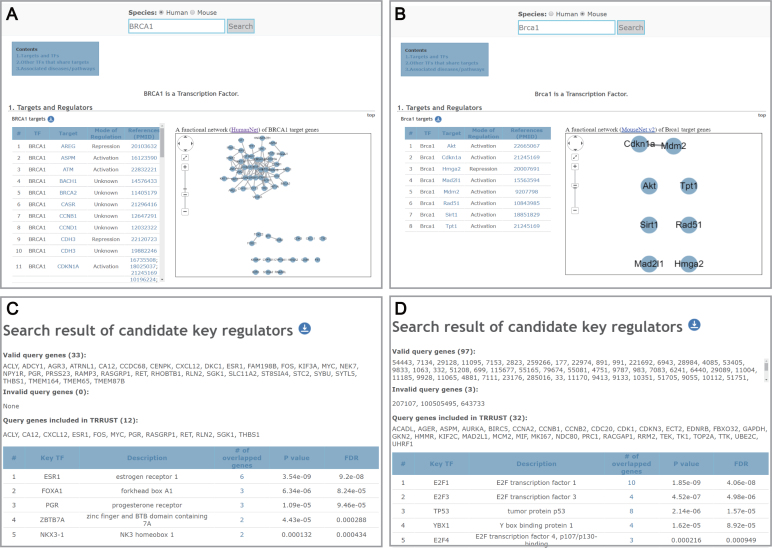
The most important improvement in TRRUST v2 over the previous version is the launch of the database of mouse TF–target interactions. The majority of mouse genes have human orthologous genes, often with the same name but with a different letter case system. Since we conducted species-specific information extraction from the literatures, TRRUST v2 generally contains different TF–target interactions between human and mouse homologous genes with the same name, as shown in the example of human BRCA1 (Figure 3A) and mouse Brca1 (Figure 3B). The TRRUST v2 web interface for a single human gene query was applied to the mouse database search with modifications for gene network visualization. If users search for targets of a particular TF, TRRUST v2 not only returns the list of regulatory targets but also returns a visualization of a network of the target genes. It will be useful to know whether the genes that are regulated by a TF are functionally associated with each other. While network visualization for human TF target genes is based on HumanNet (10) (Figure 3A), that for mouse TF target genes is based on MouseNet v2 (11) (Figure 3B). TRRUST v2 also returns a list of TFs that regulate the given query gene and the list of other TFs that significantly share the target genes. Network links among the TFs are based on protein–protein interactions derived from iRefIndex 14.0 (12). In addition to the database query, all the TF–target interactions for both humans and mice are freely available as bulk downloadable files under a Creative Commons Attribution-ShareAlike 4.0 International license (https://creativecommons.org/licenses/by-sa/4.0/).
Figure 3.
Screenshots of the search result pages for human BRCA1 gene (A) and for mouse Brca1 gene (B). Screenshots of the search result page for candidate key transcription factors (TFs) based on 33 genes responsive to siRNA knockdown of ESR1 gene (C) and based on 100 differentially expressed genes in lung tumor samples (D). In the case of lung tumor samples, only 97 of the 100 user-input genes are valid genes, which are annotated by CCDS.
Web-based prediction of key TFs
To increase the utility of TRRUST v2 database, we have newly developed a web-based prediction tool to study the transcriptional regulatory circuitry in a physiological condition. We hypothesized that if there is a statistically significant overlap between the target genes of a TF and transcriptionally responsive genes, the TF is highly likely to be involved in transcriptional regulation of the physiological condition. Therefore, we implemented the TRRUST v2 web server to search for key TFs by using Fisher’s exact test of overlap between the target genes of each TF and the genes submitted by users. To reduce false positives, we excluded TFs with less than five target genes from the statistical test and TFs with only a single overlapped gene. False discovery rate for each overlap was also calculated with justification using the Benjamini–Hochberg procedure. The TFs with significant P-values are highly likely to be key regulators of the physiological condition depicted by the responsive genes.
Next, to verify the prediction power of this method, we used a list of genes that were differentially expressed upon knockdown of a TF. Previously, Muthukaruppan et al. (13) reported 50 microarray probes corresponding to 33 human genes with differential expression under siRNA knockdown of the ESR1 gene in human MCF7 cells. We prioritized human TFs by significance of overlap between their target genes and these 33 genes, and could correctly retrieve the perturbed gene, ESR1, as the top candidate TF (Figure 3C). We then tested the feasibility of identifying disease-associated TFs using TRRUST v2. To the end, we compiled gene expression data for 46 non-small cell lung cancer (NSCLC) samples and 45 matched normal samples from GSE18842 of Gene Expression Omnibus database (14). Using the LIMMA package, we then selected the top 100 most significantly differentially expressed genes in tumor samples and submitted them to the TRRUST v2 web server to prioritize key TFs for NSCLC. E2F1, E2F3 and TP53 appeared as the top candidates with significant P-values (1.85e-9, 4.52e-7 and 2.14e-6, respectively) (Figure 3D). We could confirm their association with NSCLC via literature examination. For example, mice lacking E2F1 have been reported to develop NSCLC (15), and E2F1 has been reported to be overexpressed in NSCLC patients and high E2F1 expression is correlated with poorer clinical outcomes (16). E2F3 also shows over-expression in human lung cancer (17), and patients with high E2F3 expression show poor survival rates (18). TP53 is mutated in more than 70% of lung cancer cases (19) and has been suggested as a therapeutic target (20). These results suggest that TRRUST v2 can facilitate the identification of disease-associated TFs based on gene expression data of disease samples.
DISCUSSION
With the addition of 6552 mouse TF–target interactions, the new version of TRRUST, TRRUST v2, is now the most comprehensive database based on manual curation for both human and mouse TF–target interactions. Therefore, TRRUST v2 can facilitate comparative analysis of transcriptional regulatory circuitries between humans and mice. In a previous study, large-scale TF footprint data for humans and mice were used to investigate the conservation of trans-acting circuitry during mammalian regulatory evolution and showed ∼20% of conservation between two species (2). We found that ∼15% of human TF–target interactions of TRRUST v2 were conserved in mice. The lower conservation of literature-based TF–target interactions could be due to the study-biases between humans and mice. Interestingly, the results of the large-scale TF footprint study suggested that conservation of cis-regulatory features is low, whereas that of trans-regulatory features such as TF-to-TF associations and regulatory network architecture is high. Notably, the authors estimated that conservation of regulatory network architecture between humans and mice is higher than 95%. It would be interesting to study the evolution of regulatory circuitry using literature-based TF–target interactions as well.
We previously demonstrated the utility of TRRUST (version 1) in benchmarking transcriptional regulatory networks inferred from high-throughput experimental data. From a more practical point of view, many users of bioinformatics resources might also want to identify highly probable candidate genes for their context of interest such as disease condition. Therefore, in TRRUST v2, we implemented a network-based algorithm to prioritize TFs for a biological context that is depicted by transcriptionally responsive genes. We also validated the web-based predictions using two case studies: correct retrieval of ESR1 using genes differentially expressed upon ESR1 knockdown, and literature validation of the top three TF candidates for lung cancer, predicted by using differentially expressed genes in tumor samples. Our results showed that TRRUST v2 not only provides information extracted from the literatures but can also generate novel functional hypotheses to study the transcriptional regulation involved in human diseases. Generation of functional hypothesis using co-functional networks have increased the popularity of such networks, and various network-based algorithms have been developed (21). Algorithms also need to be developed based on regulatory gene networks to identify key transcriptional regulators associated with human diseases.
TRRUST v2 also has many TF–target interactions that are not included in other databases. Since double-checks were performed by experienced curators for most interactions, we believe that the complementary interactions in TRRUST v2 are mostly true. We also observed many TF–target interactions that are not present in TRRUST v2 but were retrieved for other databases. Assuming that all the databases compared in this study were constructed by manual curation, these non-overlaps are more likely complementary interactions rather than errors. This relative low concordance can be attributed to the difference in sensitivity between text mining methods. Then, it would also be worth to consolidate regulatory interactions from different databases to create a single more comprehensive database.
Conducting manual curation using entire candidate sentences would be a formidable task. Therefore, for more cost-effective manual curation, we prioritized the candidate sentences for harboring information regarding TF–target interactions and showed that the higher ranked sentences were more likely to be true positives. We conducted manual curation for only the top ∼28 000 and ∼24 000 sentences for humans and mice, respectively. However, continued manual curation for the remaining candidate sentences will increase the size of the database, although the retrieval rate would keep decreasing as we move to lower ranked sentences. For example, the retrieval rate of mouse TF–target interactions substantially decreased after the top 15 000 sentences, but the lower ranked ones still have a 10% or higher chance to contain information regarding TF–target interactions. Furthermore, the probability of true-positive sentences could be improved by developing new prioritizing schemes. Moreover, the upcoming published articles will continue to contribute database improvement.
In conclusion, we believe that TRRUST v2 is a versatile and expandable database of TF–target interactions, which will be a useful bioinformatics resource for the study of transcriptional regulatory circuitry in human diseases.
FUNDING
National Research Foundation of Korea [2015R1A2A1A15055859, 2017M3A9B4042581]; Brain Korea 21 (BK21) PLUS program (to I.L.). Funding for open access charge: National Research Foundation of Korea.
Conflict of interest statement. None declared.
REFERENCES
- 1. Han H., Shim H., Shin D., Shim J.E., Ko Y., Shin J., Kim H., Cho A., Kim E., Lee T. et al. TRRUST: a reference database of human transcriptional regulatory interactions. Sci. Rep. 2015; 5:11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stergachis A.B., Neph S., Sandstrom R., Haugen E., Reynolds A.P., Zhang M., Byron R., Canfield T., Stelhing-Sun S., Lee K. et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014; 515:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ravasi T., Suzuki H., Cannistraci C.V., Katayama S., Bajic V.B., Tan K., Akalin A., Schmeier S., Kanamori-Katayama M., Bertin N. et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010; 140:744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coordinators N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016; 44:D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Farrell C.M., O’Leary N.A., Harte R.A., Loveland J.E., Wilming L.G., Wallin C., Diekhans M., Barrell D., Searle S.M., Aken B. et al. Current status and new features of the Consensus Coding Sequence database. Nucleic Acids Res. 2014; 42:D865–D872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Portales-Casamar E., Kirov S., Lim J., Lithwick S., Swanson M.I., Ticoll A., Snoddy J., Wasserman W.W.. PAZAR: a framework for collection and dissemination of cis-regulatory sequence annotation. Genome Biol. 2007; 8:R207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Essaghir A., Toffalini F., Knoops L., Kallin A., van Helden J., Demoulin J.B.. Transcription factor regulation can be accurately predicted from the presence of target gene signatures in microarray gene expression data. Nucleic Acids Res. 2010; 38:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao F., Xuan Z., Liu L., Zhang M.Q.. TRED: a Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Res. 2005; 33:D103–D107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yusuf D., Butland S.L., Swanson M.I., Bolotin E., Ticoll A., Cheung W.A., Zhang X.Y., Dickman C.T., Fulton D.L., Lim J.S. et al. The transcription factor encyclopedia. Genome Biol. 2012; 13:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee I., Blom U.M., Wang P.I., Shim J.E., Marcotte E.M.. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011; 21:1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim E., Hwang S., Kim H., Shim H., Kang B., Yang S., Shim J.H., Shin S.Y., Marcotte E.M., Lee I.. MouseNet v2: a database of gene networks for studying the laboratory mouse and eight other model vertebrates. Nucleic Acids Res. 2016; 44:D848–D854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Razick S., Magklaras G., Donaldson I.M.. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008; 9:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muthukaruppan A., Lasham A., Woad K.J., Black M.A., Blenkiron C., Miller L.D., Harris G., McCarthy N., Findlay M.P., Shelling A.N. et al. Multimodal assessment of estrogen receptor mRNA profiles to quantify estrogen pathway activity in breast tumors. Clin. Breast Cancer. 2017; 17:139–153. [DOI] [PubMed] [Google Scholar]
- 14. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamasaki L., Jacks T., Bronson R., Goillot E., Harlow E., Dyson N.J.. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996; 85:537–548. [DOI] [PubMed] [Google Scholar]
- 16. Huang C.L., Liu D., Nakano J., Yokomise H., Ueno M., Kadota K., Wada H.. E2F1 overexpression correlates with thymidylate synthase and survivin gene expressions and tumor proliferation in non small-cell lung cancer. Clin Cancer Res. 2007; 13:6938–6946. [DOI] [PubMed] [Google Scholar]
- 17. Cooper C.S., Nicholson A.G., Foster C., Dodson A., Edwards S., Fletcher A., Roe T., Clark J., Joshi A., Norman A. et al. Nuclear overexpression of the E2F3 transcription factor in human lung cancer. Lung Cancer. 2006; 54:155–162. [DOI] [PubMed] [Google Scholar]
- 18. Chou Y.T., Hsieh C.H., Chiou S.H., Hsu C.F., Kao Y.R., Lee C.C., Chung C.H., Wang Y.H., Hsu H.S., Pang S.T. et al. CITED2 functions as a molecular switch of cytokine-induced proliferation and quiescence. Cell Death Differ. 2012; 19:2015–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mogi A., Kuwano H.. TP53 mutations in nonsmall cell lung cancer. J. Biomed. Biotechnol. 2011; 2011:583929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rousseau B., Jacquot C., Le Palabe J., Malleter M., Tomasoni C., Boutard T., Sakanyan V., Roussakis C.. TP53 transcription factor for the NEDD9/HEF1/Cas-L gene: potential targets in non-small cell lung cancer treatment. Sci. Rep. 2015; 5:10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shim J.E., Lee T., Lee I.. From sequencing data to gene functions: co-functional network approaches. Anim. Cells Syst. 2017; 21:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]