Abstract
The BIG Data Center at Beijing Institute of Genomics (BIG) of the Chinese Academy of Sciences provides freely open access to a suite of database resources in support of worldwide research activities in both academia and industry. With the vast amounts of omics data generated at ever-greater scales and rates, the BIG Data Center is continually expanding, updating and enriching its core database resources through big-data integration and value-added curation, including BioCode (a repository archiving bioinformatics tool codes), BioProject (a biological project library), BioSample (a biological sample library), Genome Sequence Archive (GSA, a data repository for archiving raw sequence reads), Genome Warehouse (GWH, a centralized resource housing genome-scale data), Genome Variation Map (GVM, a public repository of genome variations), Gene Expression Nebulas (GEN, a database of gene expression profiles based on RNA-Seq data), Methylation Bank (MethBank, an integrated databank of DNA methylomes), and Science Wikis (a series of biological knowledge wikis for community annotations). In addition, three featured web services are provided, viz., BIG Search (search as a service; a scalable inter-domain text search engine), BIG SSO (single sign-on as a service; a user access control system to gain access to multiple independent systems with a single ID and password) and Gsub (submission as a service; a unified submission service for all relevant resources). All of these resources are publicly accessible through the home page of the BIG Data Center at http://bigd.big.ac.cn.
INTRODUCTION
The BIG Data Center (http://bigd.big.ac.cn) at Beijing Institute of Genomics (BIG) of the Chinese Academy of Sciences (CAS) was officially founded in 2016, with the aim to provide freely open access to a suite of database resources in support of worldwide research activities in both academia and industry (1). With the increasing capability in high-throughput genome sequencing, research projects based on sequencing a variety of creatures (e.g. Earth BioGenome Project) that range from humans (e.g. US Precision Medicine Initiative (2), UK10K Project (3), China Precision Medicine Projects (4,5)) to animals (e.g. Dog 10K Genomes Project; http://dog10kgenomes.org) to plants (e.g. Arabidopsis 1001 Genomes Project; http://1001genomes.org) to microorganisms (e.g. National Microbiome Initiative (6)), are ongoing or in the planning stages around the world, consequently leading to huge amounts of omics data that are generated at ever-greater scales and rates. As a corollary, the BIG Data Center, in close collaboration with partner institutions, is continually expanding, updating and enriching its database resources through big-data integration and value-added curation, with significant improvements and advances over the previous version. Here we provide a summary of new developments and recent updates and describe core database resources of the BIG Data Center (Figure 1). All resources described are available at http://bigd.big.ac.cn and the data underlying these resources are publicly accessible for download at ftp://download.big.ac.cn.
Figure 1.
The BIG Data Center's core data resources. A full list of data resources, which contains links to each resource, is available at http://bigd.big.ac.cn/databases.
NEW DEVELOPMENTS
BioProject
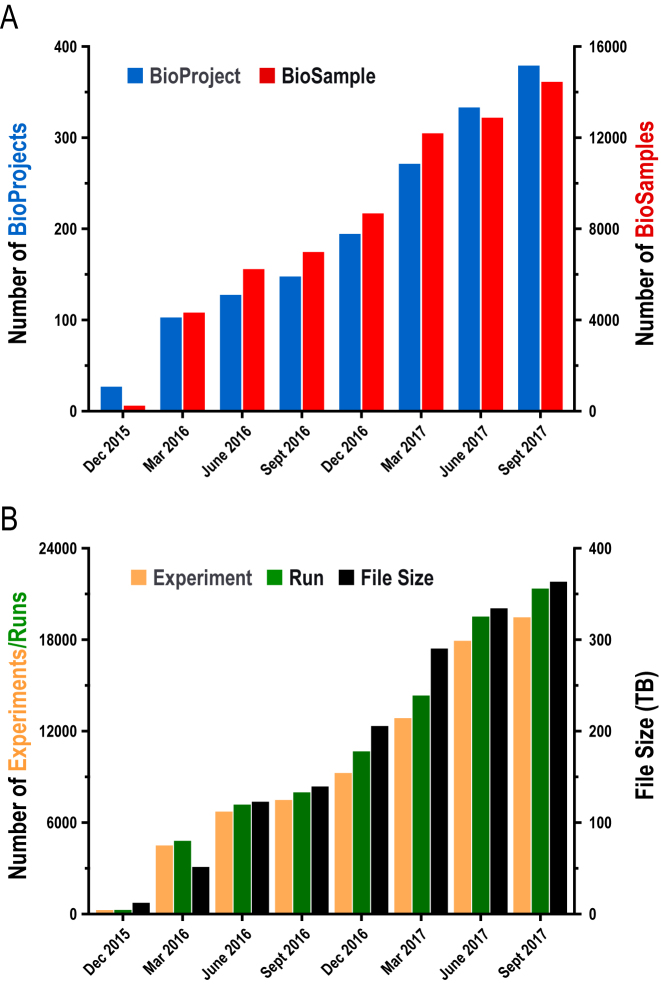
The BioProject database (http://bigd.big.ac.cn/bioproject), designed in compliance with the INSDC (International Nucleotide Sequence Database Collaboration) standards, serves as an organizational framework to provide a centralized access to descriptive metadata about research projects, ranging from genomic, transcriptomic, epigenomic and metagenomic sequencing efforts to genome-wide association studies and variation analyses. Typically, a BioProject record contains indispensable metadata information about project description, organism, data type, submitter name and affiliation, release date, grant and publication(s) if available, therefore enabling project information to be shared consistently among all relevant resources (e.g. BioSample, Genome Sequence Archive) in the BIG Data Center. BioProject also supports umbrella project, which is very useful to function as an organizational structure for any large project that is often composed of multiple sub-projects working collaboratively under a same grant. In addition, BioProject offers friendly and intuitive web interfaces for data browse and retrieval and accepts data submissions from all over the world. Till September 2017, BioProject houses a total of 380 projects submitted by 215 users from 72 organizations, presenting a dramatic increase in data submission in the past more than one year (Figure 2A).
Figure 2.
Statistics of data submissions to BioProject, BioSample and GSA. (A) Data statistics of BioProject and BioSample. (B) Data statistics of Experiments and Runs as well as file size in GSA. All statistics are frequently updated and publicly available at http://bigd.big.ac.cn/bioproject/, http://bigd.big.ac.cn/biosample/ and http://bigd.big.ac.cn/gsa/.
BioSample
The BioSample Database (http://bigd.big.ac.cn/biosample), also designed under the INSDC standards, provides structured and indexed descriptive information on biological samples. A BioSample record often contains crucial information about biological materials used in the experiments, including sample types and attributes, therefore providing basic context to the derived data. BioSample also provides reciprocal links to BioProject as well as other relevant database resources, facilitating sample search in different databases. Similarly, BioSample also accepts submissions from all over the world and as of September 2017 has accommodated a total of 14,453 samples for more than 120 species (Figure 2A).
BioCode
BioCode (http://bigd.big.ac.cn/biocode) is a centralized repository archiving bioinformatics tool codes for open source projects. It not only hosts a wide range of bioinformatics codes developed for different data analysis purposes but also collects a variety of metadata information for each tool, such as tool name, description, category, publication(s), citation count, contact information of tool owner, and organization. As of September 2017, BioCode houses a total of 6977 bioinformatics tools that are integrated through automated literature mining from highly-profile journals in the field of bioinformatics (such as Bioinformatics, BMC Bioinformatics, Genome Biology and Nucleic Acids Research). In addition, BioCode allows any user to submit tools and thus can function as an archival hub for bioinformatics tool owners to host tool codes, software packages, associated documentation and other relevant metadata information. Thus, BioCode facilitates community efforts in archiving bioinformatics tools in a centralized manner, accordingly making all bioinformatics tools publicly accessible and searchable. Together, BioCode serves as an open content platform not only for tool owners to host, archive and release bioinformatics tools but also for tool users to effectively browse, search and download any tool of interest.
ICG
ICG (http://icg.big.ac.cn) is a wiki-based, publicly editable and open-content resource for community curation of internal control genes across a diversity of species. As quantitative reverse transcription PCR (RT-qPCR) is widely used for quantification of gene expression levels (7,8), appropriate selection of internal control genes is fundamentally essential for reliable RT-qPCR normalization and accurate expression profiling (9). Toward this end, ICG is dedicated to integrating experimentally validated internal control genes from published literature and making these genes and their associated experimental conditions well-organized and public accessible to the whole scientific community. The current implementation of ICG houses >750 internal control genes covering 73 animals, 115 plants, 12 fungi and 9 bacteria. Additionally, it also incorporates detailed information on recommended application scenarios corresponding to specific experimental conditions, which, collectively, are very useful for users to adopt appropriate internal control genes for their own experiments. Thus, ICG serves as a publicly editable and open-content encyclopedia of internal control genes and accordingly bears broad utility for reliable RT-qPCR normalization and gene expression characterization in both model and non-model organisms.
BIG Search
BIG Search is a scalable text search engine that provides an inter-domain navigation and uniform access to a variety of database resources. Technically, BIG Search consists of Data Provider, Data Indexer, Distributed Search Engine and Search Interface. In the current release, the Data Provider includes not only all database resources hosted at the BIG Data Center but also seven partner databases (http://bigd.big.ac.cn/partners), viz., AnimalTFDB (10) that is a comprehensive resource for animal transcription factors, dbPAF (11) that is a resource of protein phosphorylation events in human, animals and fungi, DEG (12) that is a database of essential genes, DoriC (13) that is a database of replication origins for prokaryotic genomes, LncRNADisease (14) that is a database of lncRNA-disease associations, lncRNASNP (15) that is a database of variants in lncRNAs and PLMD (16) that is an integrative resource of 20 types of protein lysine modifications, which together make different types of data searchable by providing an index file in JSON format. The Data Indexer, including batch indexer and real-time indexer, is developed for large-scale initial data importing and small-scale real-time data updating, respectively. The Distributed Search Engine that is based on ElasticSearch (https://www.elastic.co; a highly scalable open-source full-text search and analytics engine based on Apache Lucene), plays a significant role throughout the whole platform and enables users to get access to powerful retrieval and analytical capabilities to all processed data by the Data Indexer. The Search Interface communicates with the distributed search engine in high-performance TCP protocol to present retrieval data through a web page. As a new feature in the BIG Data Center, BIG Search hosts a vast amount of data indexes from multiple resources and provides powerful text search functionality, enabling users to have inter-domain navigation and access to a wide range of biological data in one place. BIG Search has been implemented as a user-friendly search service and is accessible at the home page of the BIG Data Center.
BIG Single Sign-On
The BIG Single Sign-On (SSO; http://bigd.big.ac.cn/sso) is a user access control system that can be used to gain access to multiple independent systems with a single ID and password. BIG SSO is developed based on the Central Authentication Service, which is an open source web protocol that permits a user to access multiple web systems while just providing their credentials only once. Currently, BIG SSO provides user authentication services for a family of database systems, enabling users to sign-on any system once and grant access to the rest of systems without using different usernames or passwords. Ongoing developments are installing SSO in other database resources of the BIG Data Center where authentication is required for data submission.
Gsub
Gsub (http://bigd.big.ac.cn/gsub) is a unified submission portal providing submission services for a variety of database resources of the BIG Data Center. Armed with the BIG SSO, Gsub facilitates users to submit data in a single place and accordingly delivers a one-stop service for data submission to BioCode, BioProject, BioSample, Genome Sequence Archive, Genome Warehouse and Genome Variation Map. Equipping with Gsub, users can retrieve a list of submission history including finished and unfinished submissons, create a new submission, or delete/edit any unfinished submission. All submitted data are under quality control to ensure all necessary information complete and publicly available, but it is the submitter's responsibility for data accuracy and reliability. In addition, Gsub not only provides friendly web interfaces for metadata collection, but also supplies an FTP server for data file uploading. Ongoing developments are deploying Gsub in other database resources of the BIG Data Center, where online submission is involved.
RECENT UPDATES
Genome Sequence Archive
The Genome Sequence Archive (GSA; http://bigd.big.ac.cn/gsa) is a data repository for archiving raw sequence reads. It accepts data submissions from all over the world and provides free access to all publicly available data for global scientific communities. In the past year, GSA has been significantly upgraded, with better architecture in data structure, improved quality control and more intuitive interfaces for data presentation. Particularly, ‘BioProject’ and ‘BioSample’ have been separated from GSA as standalone databases (as described above), which serve as uniform access points for all database resources if needed. The current implementation of GSA dedicated for collecting ‘Experiment’ and ‘Run’, presents better architecture in data structure, and accordingly, along with BioProject and BioSample, is more effective in data management and exchange. In addition, GSA is enhanced by improving data curation process as well as automated quality control mechanisms and also by accepting more types of sequencing data, like PacBio RS and Complete Genomics native. As of September 2017, GSA archives a total of 19,484 Experiments and 21,363 Runs and houses more than 360 Terabytes of sequencing data in size (Figure 2B). All released data in GSA are publicly available through the FTP site at ftp://download.big.ac.cn/gsa/.
Genome Warehouse
The Genome Warehouse (GWH; http://bigd.big.ac.cn/gwh) is a public repository housing genome-scale data for a wide range of species and providing freely open access to all public available genomes. Compared to the previous release, GWH is not only enriched by integrating 138 newly released genomes (61 animals and 77 plants) from NCBI (17) and sequenced in-house (e.g. rubber tree (18)), but also significantly upgraded by developing a series of web services for genome data submission, release and sharing. Particularly, the updated implementation of GWH is able to accept data submission from all over the world and offer standardized quality control for genome sequence and genome annotation. Since the availability of submission service online in July 2017, GWH has accommodated 3 genome submissions, viz., two animals and one plant, showing the great promise to have more and more genome data submissions in the wake of high-throughput sequencing capability and large-scale sequencing-based projects as mentioned above. For each collected species, GWH incorporates detailed descriptive information including biological sample metadata, genome assembly metadata, sequence data and genome annotation. Future directions of GWH include continuous integration of newly sequenced genomes, improvement of genome data quality control and genome annotation, and development of more friendly and interactive interfaces for data presentation and visualization.
Genome Variation Map
The Genome Variation Map (GVM; http://bigd.big.ac.cn/gvm), is a public data repository of genome variations. GVM aims to collect, integrate and visualize genome variations for a wide range of species, accepts submissions of different types of genome variations from all over the world and provides free open access to all publicly available data in support of worldwide research activities. By comparison with the previous version, the current implementation of GVM houses a total of ∼4.9 billion variants for 19 species, with particular focuses on human, cultivated plants (e.g. maize, rice, tomato, sorghum, soybean), domesticated animals (e.g. chicken, dog, goat, pig) and featured species (e.g. giant panda, killer whale, moso bamboo, rubber, wheat). In addition, based on manual curation from a number of publications, GVM incorporates 8,669 individual genotypes and 13,262 high-quality genotype-to-phenotype (G2P) associations for non-human species, and also integrates from ClinVar (19), GWAS-catalog (20) and OMIM (21) a comprehensive collection of 180,911 G2P pairs for human. Moreover, it is significantly improved by developing intuitive web interfaces for data submission, browse and search and deploying an interactive user-friendly genome browser for visualization of variant genotype and allele frequency. Therefore, GVM serves as an important resource for archiving genomic variation data, helpful for better understanding population genetic diversity and deciphering complex mechanisms associated with different phenotypes.
Gene Expression Nebulas
The Gene Expression Nebulas (GEN; http://bigd.big.ac.cn/gen) is a data portal of gene expression profiles under various conditions derived entirely from RNA-Seq data analysis in multiple species. Compared to the previous release, GEN hosts three featured resources, namely, Mammalian Transcriptomic Database (22), Rice Expression Database (23,24) and ICG that is newly developed as a knowledgebase of internal control genes for RT-qPCR normalization (as described above). Since ICG provides empirical candidates for further evaluation of dynamic expression profiles under diverse experimental conditions, internal control genes are valuable references for accurate gene expression normalization in GEN. Therefore, ongoing efforts are primarily paid to not only integration of high-quality RNA-Seq data from featured species with high-quality genome sequences and annotation information but also development of methods for accurate gene expression profiling based on ICG. Equally important, as RNA-Seq raw data are stored in GSA but their analyzed results are stored in GEN, we also plan to bridge the gap between GSA and GEN by building standardized pipelines for RNA-Seq data analysis and deploying these pipelines into a cloud platform, where raw data in GSA can be automatically analyzed and the corresponding results can be automatically exported into GEN.
Methylation Bank
The Methylation Bank (MethBank; http://bigd.big.ac.cn/methbank) (25) is a methylation databank focusing on health and aging of humans, embryonic development of animals and growth and development of plants. Compared to the previous version, MethBank is enriched by accommodating a larger number of genome-wide high-quality DNA methylomes from multiple species. Specifically, it integrates 34 methylomes from 4,577 peripheral blood samples of healthy people at different ages, 336 from different developmental stages and/or tissues in five economical important plants, and 18 from gametes and early embryos at multiple stages in two animals. Moreover, it is enhanced by improving the functionalities for data annotation, leading to identification of methylation sites closely associated with age, sites with constant methylation levels across different ages, age-specific differentially methylated cytosines/regions, differentially methylated promoters, and methylated CpG islands. Also, MethBank equips with more friendly web interfaces to retrieve a diversity of methylation-related information for a specific gene or a genomic region and provides two tools to facilitate online estimation of human age based on methylation sites and to identify differentially methylated promoters via Fisher's exact test and FDR correction, respectively. Future directions are frequent integration of more high-quality methylomes from a wider range of organisms, improvement of functionalities for data annotation and presentation and development of new functionalities for methylation data submission.
Science Wikis
Science Wikis (http://bigd.big.ac.cn/sciencewikis) is a series of biological knowledge wikis that are built based on wiki technology or wiki concept to harness collective intelligence in community curation—allowing any user to create/edit any content, accordingly featuring community-contributed contents, low cost for maintenance and broader content coverage. The current release of Science Wikis consists of five wiki-based databases (including LncRNAWiki (26), RiceWiki (27), ESND (28), WikiCell (29), and one new database, ICG) as well as several wiki extensions that achieve customized functionalities, such as AuthorReward (30) that quantifies users contribution in biological wikis and provides explicit authorship as a reward. Over the past year, the major updates of Science Wikis are as follows. LncRNAWiki (http://lncrna.big.ac.cn) is updated by curating more long non-coding RNAs (lncRNAs) and also associating lncRNAs with human diseases; a total of 959 human lncRNAs have been manually community-curated based on published literatures and 438 of them have been experimentally validated to be associated with cancer and other diseases. RiceWiki (http://ricewiki.big.ac.cn) is updated by integrating ∼120 rice genes’ annotations from recent publications and the current version contains ∼500 manually community-curated rice genes. ICG, as mentioned above, is a knowledgebase of internal control genes for RT-qPCR normalization, integrating more than 750 internal control genes curated from a large volume of literatures and thus aiding users to choose appropriate internal control genes corresponding to specific experimental conditions for both model and non-model organisms. Ongoing developments are integrating BIG SSO into all associated databases of Science Wikis, which can ease users to get involved in community curation by sign-in only once and also provide a summary of community-curated contributions to multiple databases.
CONCLUDING REMARKS
The BIG Data Center delivers a suite of database resources in support of research activities in both academia and industry. Considering the increasing quantities of biological data generated by large-scale sequencing projects around the world, the BIG Data Center is committed to integrating a wide range of biological data, developing a variety of database resources and services, and conducting basic research in aid of transformation of big data into big discoveries. The BIG Data Center, with the increasing funding support from CAS and the government, will definitely continue to grow to become indispensable for worldwide research activities by working collaboratively with partner institutions to address critical challenges in biological big data deposition, integration and translation.
ACKNOWLEDGEMENTS
We thank a number of users for submitting data, providing annotations, sending suggestions and reporting bugs. The BIG Data Center is indebted to its funders, including the Chinese Academy of Sciences, the Ministry of Science and Technology of China, the Natural Science Foundation of China, and Beijing Institute of Genomics.
APPENDIX
Corresponding author: Zhang Zhang1,2,3,4,*
Co-corresponding authors: Wenming Zhao1,3,4,*, Jingfa Xiao1,2,3,4,*, Yiming Bao1,2,3,*
BIG DATA CENTER MEMBERS (Arranged by project role and then by contribution except for Team Leader, as indicated; DCA = Data Curation & Analysis; DSD = Database System Development; TL = Team Leader)
BioCode Team: DSD: Xingjian Xu1,2,3,a,#; TL: Lili Hao1,2,#
BioProject, BioSample & GSA Team: DSD: Junwei Zhu1,#, Bixia Tang1,2,3, Qing Zhou1,3, Fuhai Song2,3; DCA: Tingting Chen1,#, Sisi Zhang1,#, Lili Dong1, Li Lan1; TL: Yanqing Wang1,#
GWH Team: DCA: Jian Sang1,2,3,#, Lili Hao1,2, Fang Liang1, Jiabao Cao1,2,3, Fang Liu5, Lin Liu1,2,3; DSD: Fan Wang1,2,#, Yingke Ma1,2, Xingjian Xu1,2,3,a, Lijuan Zhang1,2,3; TL: Meili Chen1,2,#
GVM Team: DCA: Dongmei Tian1,#, Cuiping Li1,#, Lili Dong1,#, Zhenglin Du1, Na Yuan1, Jingyao Zeng1, Zhewen Zhang1,2, Jinyue Wang1,2,3, Shuo Shi1,2,3, Yadong Zhang1,2,3, Mengyu Pan1,2,3; DSD: Bixia Tang1,2,3,#, Dong Zou1,2; TL: Shuhui Song1,#
GEN Team: DCA: Jian Sang1,2,3,#, Lin Xia1,2,3,#, Zhennan Wang3,6, Man Li1,2,3, Jiabao Cao1,2,3, Guangyi Niu1,2,3, Yang Zhang1,2,3, Xin Sheng1,2,3, Mingming Lu1,2,3, Qi Wang1,2,3, Jingfa Xiao1,2,3,4,*; DSD: Dong Zou1,2,#, Fan Wang1,2,3; TL: Lili Hao1,2,#
MethBank Team: DCA: Fang Liang1,#, Mengwei Li1,2,3,#, Shixiang Sun1,2,3,b; DSD: Dong Zou1,2,#; TL: Rujiao Li1,#
Science Wikis Team: DCA: Chunlei Yu1,2,3,#, Guangyu Wang1,2,3,c,#, Jian Sang1,2,3, Lin Liu1,2,3, Mengwei Li1,2,3, Man Li1,2,3, Guangyi Niu1,2,3, Jiabao Cao1,2,3, Shixiang Sun1,2,3,b, Lin Xia1,2,3, Hongyan Yin1,2,3,d; DSD: Dong Zou1,2, Xingjian Xu1,2,3,a; TL: Lina Ma1,2,#
Hardware & System Administration Team: Huanxin Chen1 (TL), Yubin Sun1, Lei Yu1, Shuang Zhai1, Mingyuan Sun1
Writing Group: Zhang Zhang1,2,3,4,*, Wenming Zhao1,3,4,*, Jingfa Xiao1,2,3,4,*, Yiming Bao1,2,3,*, Shuhui Song1, Lili Hao1,2, Rujiao Li1, Lina Ma1,2, Jian Sang1,2,3, Yanqing Wang1, Bixia Tang1,2,3, Dong Zou1,2, Fan Wang1,2
BIG DATA CENTER PARTNERS (Listed in alphabetical order by database resources)
AnimalTFDB & lncRNASNP Team: Ya-Ru Miao7, An-Yuan Guo7
dbPAF Team: Shaofeng Lin7, Yu Xue7
DEG & DoriC Team: Hao Luo8,9,10, Feng Gao8,9,10
LncRNADisease Team: Wei Ma11,12, Qinghua Cui11
PLMD Team: Haodong Xu7, Yu Xue7
1BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China
2CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China
3University of Chinese Academy of Sciences, Beijing 100049, China
4Collaborative Innovation Center of Genetics and Development, Fudan University, Shanghai 200438, China
5College of Computer Science Technology, Inner Mongolia Normal University, Hohhot, Inner Mongolia 010010, China
6State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China
7Key Laboratory of Molecular Biophysics of Ministry of Education, College of Life Science and Technology and the Collaborative Innovation Center for Biomedical Engineering, Huazhong University of Science and Technology, Wuhan, Hubei 430074, China
8Department of Physics, Tianjin University, Tianjin 300072, China
9Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin 300072, China
10SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin University, Tianjin 300072, China
11Department of Biomedical Informatics, MOE Key Laboratory of Cardiovascular Sciences, School of Basic Medical Sciences, Peking University, Beijing 100191, China
12Central Laboratory, Navy General Hospital of PLA, Beijing 100048, China
aPresent address: College of Computer Science Technology, Inner Mongolia Normal University, Hohhot, Inner Mongolia 010010, China
bPresent address: Department of Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA
cPresent address: Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030, USA
dPresent address: Hainan Key Laboratory for Sustainable Utilization of Tropical Bioresources, Institute of Tropical Agriculture and Forestry, Hainan University, Haikou, Hainan 570228, China
*To whom correspondence should be addressed: Zhang Zhang (zhangzhang@big.ac.cn). Correspondence may also be addressed to Wenming Zhao (zhaowm@big.ac.cn), Jingfa Xiao (xiaojingfa@big.ac.cn), and Yiming Bao (baoym@big.ac.cn).
#The authors wish it to be known that, in their opinion, these authors should be regarded as Joint First Authors.
Contributor Information
BIG Data Center Members:
Xingjian Xu, Lili Hao, Junwei Zhu, Bixia Tang, Qing Zhou, Fuhai Song, Tingting Chen, Sisi Zhang, Lili Dong, Li Lan, Yanqing Wang, Jian Sang, Lili Hao, Fang Liang, Jiabao Cao, Fang Liu, Lin Liu, Fan Wang, Yingke Ma, Xingjian Xu, Lijuan Zhang, Meili Chen, Dongmei Tian, Cuiping Li, Lili Dong, Zhenglin Du, Na Yuan, Jingyao Zeng, Zhewen Zhang, Jinyue Wang, Shuo Shi, Yadong Zhang, Mengyu Pan, Bixia Tang, Dong Zou, Shuhui Song, Jian Sang, Lin Xia, Zhennan Wang, Man Li, Jiabao Cao, Guangyi Niu, Yang Zhang, Xin Sheng, Mingming Lu, Qi Wang, Jingfa Xiao, Dong Zou, Fan Wang, Lili Hao, Fang Liang, Mengwei Li, Shixiang Sun, Dong Zou, Rujiao Li, Chunlei Yu, Guangyu Wang, Jian Sang, Lin Liu, Mengwei Li, Man Li, Guangyi Niu, Jiabao Cao, Shixiang Sun, Lin Xia, Hongyan Yin, Dong Zou, Xingjian Xu, Lina Ma, Huanxin Chen, Yubin Sun, Lei Yu, Shuang Zhai, Mingyuan Sun, Zhang Zhang, Wenming Zhao, Jingfa Xiao, Yiming Bao, Shuhui Song, Lili Hao, Rujiao Li, Lina Ma, Jian Sang, Yanqing Wang, Bixia Tang, Dong Zou, and Fan Wang
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB13040500 to W.Z. and Z.Z.; XDA08020102 to Z.Z.]; National Key Research Program of China [2017YFC0907502 to Z.Z.; 2016YFC0901603 to W.Z.; 2017YFC0907503, 2016YFB0201702, 2016YFC0901903 to J.X.]; National Programs for High Technology Research and Development [863 Program; 2015AA020108 to Z.Z.; 2015AA020101 to F.G.]; National Natural Science Foundation of China [31100915 to L.H.; 31200978 to L.M.; 31771465 and 31471248 to J.X.; 31671360 to Y.X.; 31571358 to F.G.]; International Partnership Program of the Chinese Academy of Sciences [153F11KYSB20160008]; Key Program of the Chinese Academy of Sciences [KJZD-EW-L14 to J.X.]; Key Technology Talent Program of the Chinese Academy of Sciences [to W.Z.]; The 100 Talent Program of the Chinese Academy of Sciences [to Y.B. and Z.Z.]; The Youth Innovation Promotion Association of the Chinese Academy of Sciences [2017141 to S.S.]; The Special Project on Precision Medicine under the National Key R&D Program [SQ2017YFSF090210 to Y.X.]. Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. BIG Data Center Members. The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 2017; 45:D18–D24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collins F.S., Varmus H.. A new initiative on precision medicine. N. Engl. J. Med. 2015; 372:793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taylor P.N., Porcu E., Chew S., Campbell P.J., Traglia M., Brown S.J., Mullin B.H., Shihab H.A., Min J., Walter K. et al. . Whole-genome sequence-based analysis of thyroid function. Nat. Commun. 2015; 6:5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cyranoski D. China embraces precision medicine on a massive scale. Nature. 2016; 529:9–10. [DOI] [PubMed] [Google Scholar]
- 5. Li H. Cancer precision medicine in China. Genomics Proteomics Bioinformatics. 2016; 14:325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouchie A. White House unveils National Microbiome Initiative. Nat. Biotechnol. 2016; 34:580. [DOI] [PubMed] [Google Scholar]
- 7. Bustin S.A., Benes V., Nolan T., Pfaffl M.W.. Quantitative real-time RT-PCR—a perspective. J. Mol. Endocrinol. 2005; 34:597–601. [DOI] [PubMed] [Google Scholar]
- 8. Mestdagh P., Van Vlierberghe P., De Weer A., Muth D., Westermann F., Speleman F., Vandesompele J.. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009; 10:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H.M., Liu T., Liu C.J., Song S., Zhang X., Liu W., Jia H., Xue Y., Guo A.Y.. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 2015; 43:D76–D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ullah S., Lin S., Xu Y., Deng W., Ma L., Zhang Y., Liu Z., Xue Y.. dbPAF: an integrative database of protein phosphorylation in animals and fungi. Scientific Rep. 2016; 6:23534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo H., Lin Y., Gao F., Zhang C.T., Zhang R.. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014; 42:D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao F., Luo H., Zhang C.T.. DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 2013; 41:D90–D93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q.. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013; 41:D983–D986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gong J., Liu W., Zhang J., Miao X., Guo A.Y.. lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2015; 43:D181–D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu H., Zhou J., Lin S., Deng W., Zhang Y., Xue Y.. PLMD: an updated data resource of protein lysine modifications. J. Genet. Genomics. 2017; 44:243–250. [DOI] [PubMed] [Google Scholar]
- 17. NCBI Resource Coordinators. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017; 45:D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang C., Yang M., Fang Y., Luo Y., Gao S., Xiao X., An Z., Zhou B., Zhang B., Tan X. et al. . The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants. 2016; 2:16073. [DOI] [PubMed] [Google Scholar]
- 19. Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. et al. . ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. et al. . The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A.. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015; 43:D789–D798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sheng X., Wu J., Sun Q., Li X., Xian F., Sun M., Fang W., Chen M., Yu J., Xiao J.. MTD: a mammalian transcriptomic database to explore gene expression and regulation. Brief. Bioinformatics. 2017; 18:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xia L., Zou D., Sang J., Xu X., Yin H., Li M., Wu S., Hu S., Hao L., Zhang Z.. Rice Expression Database (RED): an integrated RNA-Seq-derived gene expression database for rice. J. Genet. Genomics. 2017; 44:235–241. [DOI] [PubMed] [Google Scholar]
- 24. Zhang Z., Hu S.N., He H., Zhang H.Y., Chen F., Zhao W.M., Xiao J.F., Chen L.L., Xue Y., Wang X.F. et al. . Information Commons for Rice (IC4R). Nucleic Acids Res. 2016; 44:D1172–D1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zou D., Sun S., Li R., Liu J., Zhang J., Zhang Z.. MethBank: a database integrating next-generation sequencing single-base-resolution DNA methylation programming data. Nucleic Acids Res. 2015; 43:D54–D58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma L.N., Li A., Zou D., Xu X.J., Xia L., Yu J., Bajic V.B., Zhang Z.. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015; 43:D187–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Z., Sang J., Ma L., Wu G., Wu H., Huang D., Zou D., Liu S., Li A., Hao L. et al. . RiceWiki: a wiki-based database for community curation of rice genes. Nucleic Acids Res. 2013; 42:D1222–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dai L., Xu C., Tian M., Sang J., Zou D., Li A., Liu G., Chen F., Wu J., Xiao J. et al. . Community intelligence in knowledge curation: an application to managing scientific nomenclature. PLoS One. 2013; 8:e56961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao D., Wu J., Zhou Y., Gong W., Xiao J., Yu J.. WikiCell: a unified resource platform for human transcriptomics research. OMICS. 2012; 16:357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dai L., Tian M., Wu J., Xiao J., Wang X., Townsend J.P., Zhang Z.. AuthorReward: increasing community curation in biological knowledge wikis through automated authorship quantification. Bioinformatics. 2013; 29:1837–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]