Abstract
MetaCyc (https://MetaCyc.org) is a comprehensive reference database of metabolic pathways and enzymes from all domains of life. It contains more than 2570 pathways derived from >54 000 publications, making it the largest curated collection of metabolic pathways. The data in MetaCyc is strictly evidence-based and richly curated, resulting in an encyclopedic reference tool for metabolism. MetaCyc is also used as a knowledge base for generating thousands of organism-specific Pathway/Genome Databases (PGDBs), which are available in the BioCyc (https://BioCyc.org) and other PGDB collections. This article provides an update on the developments in MetaCyc during the past two years, including the expansion of data and addition of new features.
INTRODUCTION
MetaCyc (https://MetaCyc.org) is a highly curated reference database of metabolism from all domains of life. It contains data about chemical compounds, reactions, enzymes and metabolic pathways that have been experimentally validated and reported in the scientific literature (1). Most data in MetaCyc concerns small molecule metabolism, although an increasing amount of macromolecular metabolism (e.g. protein modification) is also present. MetaCyc is a uniquely valuable resource due to its exclusively experimentally determined data, intensive curation, extensive referencing, and user-friendly and highly integrated interface. It is commonly used in various fields, including biochemistry, enzymology, metabolomics, genome and metagenome analysis, and metabolic engineering.
In addition to its role as a general reference on metabolism, MetaCyc can be used by the PathoLogic component of the Pathway Tools software (2,3) as a reference database to computationally predict the metabolic network of any organism that has a sequenced and annotated genome (4). During this partially automated process, the predicted metabolic network is captured in the form of a Pathway/Genome Database (PGDB). Pathway Tools also provides editing tools that enable improving and updating these computationally generated PGDBs by manual curation. SRI has used MetaCyc to create almost 11 000 PGDBs (as of August 2017), which are available through the BioCyc (https://BioCyc.org) website (5). In addition, many groups outside SRI have generated thousands of additional PGDBs (6–10). Some of these groups have further improved those databases by performing their own curation. Interested scientists may adopt any of the SRI PGDBs through the BioCyc website for further curation (https://biocyc.org/BioCycUserGuide.shtml#node_sec_6).
EXPANSION OF METACYC DATA
Since the last Nucleic Acids Research publication (two years ago) (1), we added 219 new base pathways (pathways comprised of reactions only, where no portion of the pathway is designated as a subpathway) and four superpathways (pathways composed of at least one base pathway plus additional reactions or pathways), and updated 112 existing pathways, for a total of 335 new and revised pathways. The total number of base pathways grew by 9%, from 2363 (version 19.1) to 2572 (version 21.1) (the total increase is <219 pathways, because some existing pathways were deleted from the database during this period). The number of enzymes in the database grew by 10%; reactions by 13%; chemical compounds by 13%; citations by 18%; and the number of referenced organisms increased by 7% (currently at 2883). See Table 1 for a list of species with >20 experimentally elucidated pathways in MetaCyc, and Table 2 for the taxonomic distribution of all MetaCyc pathways.
Table 1. List of species with 20 or more experimentally elucidated pathways represented in MetaCyc (meaning experimental evidence exists for the occurrence of these pathways in the organism).
| Bacteria | Eukarya | Archaea | |||
|---|---|---|---|---|---|
| Escherichia coli | 343 | Arabidopsis thaliana | 337 | Methanocaldococcus jannaschii | 29 |
| Pseudomonas aeruginosa | 75 | Homo sapiens | 294 | Methanosarcina barkeri | 24 |
| Bacillus subtilis | 61 | Saccharomyces cerevisiae | 199 | Sulfolobus solfataricus | 21 |
| Pseudomonas putida | 50 | Rattus norvegicus | 84 | Methanosarcina thermophila | 20 |
| Mycobacterium tuberculosis | 45 | Glycine max | 63 | ||
| Salmonella typhimurium | 44 | Mus musculus | 56 | ||
| Pseudomonas fluorescens | 32 | Pisum sativum | 53 | ||
| Synechocystis sp. PCC 6803 | 30 | Nicotiana tabacum | 52 | ||
| Klebsiella pneumoniae | 29 | Oryza sativa | 48 | ||
| Enterobacter aerogenes | 26 | Zea mays | 47 | ||
| Agrobacterium tumefaciens | 25 | Solanum tuberosum | 43 | ||
| Mycobacterium smegmatis | 23 | Catharanthus roseus | 30 | ||
| Corynebacterium glutamicum | 21 | Spinacia oleraca | 29 | ||
| Hordeum vulgare | 26 | ||||
| Triticum aestivum | 26 | ||||
| Bos taurus | 24 | ||||
| Petunia x hybrida | 21 | ||||
| Sus scrofa | 20 |
The species are grouped by taxonomic domain and are ordered within each domain based on the number of pathways (number following species name) to which the given species was assigned.
Table 2. The distribution of pathways in MetaCyc based on the taxonomic classification of associated species.
| Bacteria | Eukarya | Archaea | |||
|---|---|---|---|---|---|
| Proteobacteria | 1181 | Viridiplantae | 986 | Euryarchaeota | 158 |
| Firmicutes | 378 | Fungi | 457 | Crenarchaeota | 42 |
| Actinobacteria | 388 | Metazoa | 401 | Thaumarchaeota | 2 |
| Cyanobacteria | 89 | Euglenozoa | 31 | ||
| Bacteroidetes/Chlorobi | 83 | Alveolata | 21 | ||
| Deinococcus-Thermus | 30 | Amoebozoa | 11 | ||
| Thermotogae | 25 | Stramenopiles | 10 | ||
| Tenericutes | 18 | Haptophyceae | 6 | ||
| Aquificae | 18 | Rhodophyta | 6 | ||
| Spirochaetes | 14 | Fornicata | 4 | ||
| Chlamydiae -Verrucomicrobia | 9 | Parabasalia | 3 | ||
| Chloroflexi | 8 | ||||
| Planctomycetes | 6 | ||||
| Fusobacteria | 6 | ||||
| Nitrospirae | 2 | ||||
| Thermodesulfobacteria | 2 | ||||
| Chrysiogenetes | 1 | ||||
| Nitrospinae | 1 |
For example, the statement ‘Tenericutes 18’ means that experimental evidence exists for the occurrence of at least 18 MetaCyc pathways in members of this taxonomic group. Major taxonomic groups are grouped by domain and are ordered within each domain based on the number of pathways (number following taxon name) associated with the taxon. A pathway may be associated with multiple organisms.
While describing in this limited space the various additions to MetaCyc data during the past two years is impossible, the following partial list of new or completely revised pathways may illustrate the breadth of topics that have been covered during this time.
Antibiotic biosynthesis. We added pathways for the biosynthesis of actinomycin D; holomycin; guadinomine B; dapdiamides; penicillin G; penicillin V; zwittermicin A; echinomycin; triostin A; ravidomycin V; indolmycin; phosalacine; tetracycline; oxytetracycline; chlorotetracycline; nocardicin A; tunicamycin; daunorubicin; mithramycin and validamycin.
Aromatic compound degradation. We added new pathways for the degradation of bisphenol A; butachlor; diphenyl ethers; resorcinol; γ-resorcylate; 1-chloro-2-nitrobenzene; 2,4-xylenol; 2,5-xylenol; 3,5-xylenol; 4-methylphenol; P-cumate; P-cymene; 4-chloronitrobenzene and pentachlorophenol.
Bacteriochlorophyll biosynthesis. We added pathways for the biosynthesis of all major forms of bacteriochlorophyll: bacteriochlorophyll a; bacteriochlorophyll b; bacteriochlorophyll c; bacteriochlorophyll d; and bacteriochlorophyll e.
Bioluminescence. We added five new pathways that describe bioluminescence in bacteria, jellyfish, corals, fireflies and dinoflagellates.
Heme degradation. We expanded our coverage of heme degradation from one to seven pathways.
Protein modification. We added pathways describing protein S-nitrosylation and denitrosylation; SAMPylation; NEDDylation; pupylation and depupylation and lipoylation. We also added pathways that describe the N-end, Ac/N-end, and Arg/N-end rules, which determine protein degradation.
Short-chain alkane and alkene degradation. New pathways were added for the degradation of butane; methyl tert-butyl ether; propane; ethane; isoprene and 2-methylpropene.
Teichoic acid biosynthesis. We added pathways for the biosynthesis of all teichoic acid forms for which metabolic knowledge exists: poly(glycerol phosphate) wall teichoic acid; poly(3-O-β-D-glucopyranosyl-N-acetylgalactosamine 1-phosphate) wall teichoic acid; poly(ribitol phosphate) wall teichoic acid (in Bacillus subtilis); poly(ribitol phosphate) wall teichoic acid (in Staphylococcus aureus); teichuronic acid; type I lipoteichoic acid and type IV lipoteichoic acid.
Mycobacterial pathways. We added several new pathways from this important human pathogen, including the biosynthesis of dimycocerosyl phthiocerol; dimycocerosyl triglycosyl phenolphthiocerol; mycobacterial sulfolipid; P-HBAD; ω-sulfo-II-dihydromenaquinone-9; phenolphthiocerol; glycogen (from α-maltose 1-phosphate) and phosphatidylinositol mannoside. We also added pathways describing isoniazid activation, ethionamide activation and protein pupylation and depupylation.
Archaeal pathways. We added new pathways that describe different mechanisms for the regeneration of the coenzyme B/coenzyme M mixed disulfide in methanogens, for the biosynthesis of factor 420 and factor 430, and for archaeal nucleoside and nucleotide degradation.
Human metabolism. New pathways describe alternative routes for the biosynthesis of the fatty acids (4Z,7Z,10Z,13Z,16Z)-docosa-4,7,10,13,16-pentaenoate, docosahexaenoate, arachidonate, and icosapentaenoate; the metabolism of bile acids and iso-bile acids; the biosynthesis and degradation of plasmalogen; the biosynthesis of the A, B, H and Lewis epitopes from both type 1 and type 2 precursor disaccharide; the modification of terminal O-glycans; the biosynthesis of i and I antigens; and the hydroxylation and glycosylation of procollagen. We have also added several pathways describing the biosynthesis of glycosphingolipids (different pathways describe the gala, ganglio, globo, lacto and neolacto series).
Plant metabolism. We performed major revisions in the areas of glucosinolate metabolism (13 new and revised pathways); jasmonic acid metabolism (four pathways); and cyanogenic glycosides biosynthesis (four pathways describing dhurrin, linamarin, lotaustralin and taxiphyllin, respectively). We also significantly revised our coverage of bitter acids biosynthesis (three pathways); pterocarpan phytoalexins biosynthesis (two pathways); camalexin biosynthesis; Amaryllidacea alkaloids biosynthesis; prunasin and amygdalin biosynthesis; anthocyanin biosynthesis and proanthocyanidins biosynthesis.
Compounds
The total number of compounds grew by 13%, from 12 362 (version 19.1) to 14 003 (version 21.1). 9442 of these compounds participate in reactions, and 13 725 have structures. Most MetaCyc compounds also contain standard Gibbs free energy of formation (ΔfG′°) values, most of which are computed by Pathway Tools using an algorithm developed internally that is based on techniques by Jankowski et al. (11) and Alberty (12). As of August 2017, a total of 13 760 compounds include these Gibbs free energy values.
Reactions
The total number of enzymatic reactions grew by 13%, from 12 701 (version 19.1) to 14 347 (version 21.1). The number of total reactions (including non-enzymatic) is 15 691. MetaCyc uses a reaction-balance-checking algorithm that checks not only for elemental composition but also for electric charge. Unlike many reaction resources available online, the vast majority of MetaCyc reactions are completely balanced, taking into account the protonation state of the compounds (which is the state most prevalent at pH 7.3). As of August 2017, MetaCyc contains 14 302 balanced reactions. The remaining 1389 reactions cannot be balanced due to assorted reasons (for example, a reaction may describe a polymeric process, such as the hydrolysis of a polymer of an undefined length, may involve an ‘n’ coefficient, or may involve a substrate that lacks a defined structure, such as ‘an aldose’).
MetaCyc reactions also contain standard change in Gibbs free energy (ΔrG′°) values that are computed based on the ΔfG′° values computed for compounds. As of August 2017, a total of 13 877 reactions include these Gibbs free energy values.
Linking to other databases
Objects in MetaCyc are extensively linked to other leading databases in the field. MetaCyc proteins have a total of 17 669 links to a number of protein databases that include (only databases with more than 1000 links are listed) InterPro; PDB; Pfam; UniProt; Protein Model Portal; PROSITE; SMR; PRIDE; PID; PANTHER; PRINTS; MODBASE; SMART; RefSeq; EcoliWiki; PortEco; DIP; MINT; ProDB; SwissModel; PhylomeDB; PhosphoSite; and CAZy. MetaCyc genes have a total of 11 860 links to NCBI-Entrez; NCBI-Gene; STRING; RegulonDB; EcoGene; EchoBase; ASAP; OU Microarray; RefSeq; MIM; CGSC; and ArrayExpress. MetaCyc compounds have a total of 14 209 links to PubChem; ChEBI; KEGG; ChemSpider; HMDB; MetaboLights; RefMet and CAS. MetaCyc reactions have a total of 15 779 links to UniProt, Rhea and KEGG.
Enzyme Commission numbers
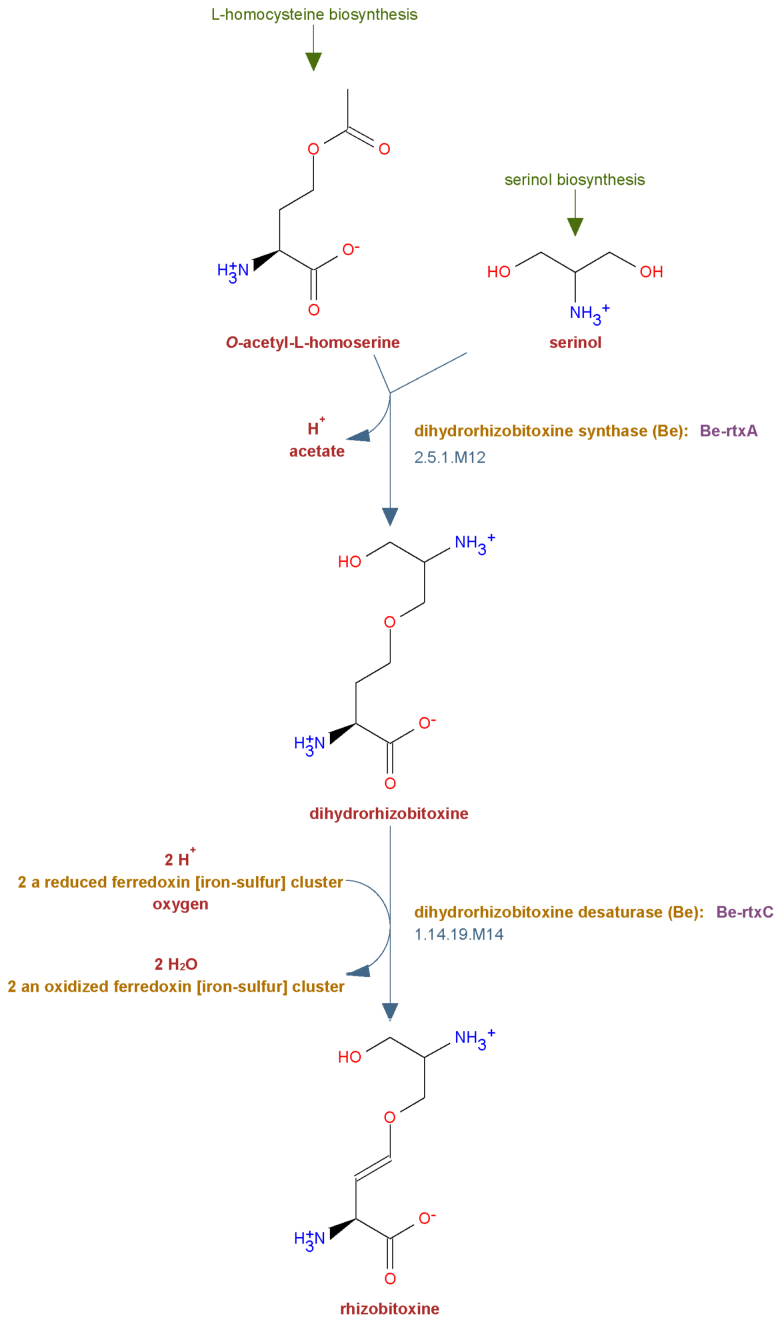
Curation of MetaCyc is conducted in close collaboration with the Enzyme Commission (EC) (13). During the curation process, MetaCyc curators come across thousands of enzymes that have not yet been classified by the EC. In addition, curation exposes errors in older existing EC entries. While curating MetaCyc content, curators prepare and submit new and revised entries to the EC, leading to the creation of hundreds of new and modified EC entries over the past two years. Many enzymes that have not been classified in the EC system are assigned ‘M-numbers’ in MetaCyc (see Figure 1), which are temporary numbers that indicate a well-characterized enzymatic activity that has not yet been classified by the EC (14). Our intention is to have as many M-numbers as possible eventually replaced by official EC numbers.
Figure 1.
A typical MetaCyc pathway. A short pathway was selected for this figure; the average number of metabolites in a MetaCyc pathway is 12.8, with the largest pathway containing 204 metabolites. The enzymes in this pathway have not yet been classified by the Enzyme Commission and were assigned M-numbers (see text). The green captions are links to the upstream pathways that produce the inputs for this pathway.
SOFTWARE AND WEBSITE ENHANCEMENTS
The following sections describe significant enhancements to Pathway Tools (the software that powers the BioCyc website) during the past two years that affect the MetaCyc user experience.
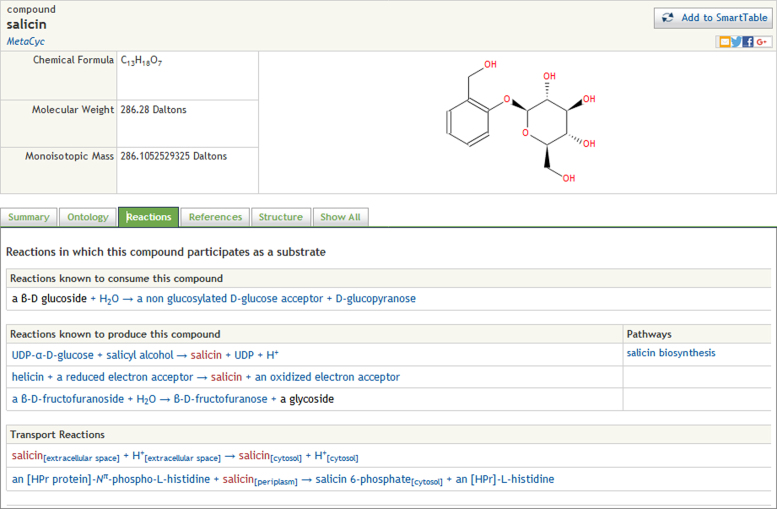
Redesigned metabolite pages
We have redesigned the Web metabolite (compound) pages to use a tabbed structure. The information shown on these pages is now divided into several tabs including a summary, ontology, reactions, and structure tabs. A ‘Show All’ tab displays all the information in one page (see Figure 2).
Figure 2.
Redesigned compound pages use a tabbed interface to reduce clutter on information pages.
Update notifications
MetaCyc has a new capability to inform users of newly curated information in specified areas of interest. The update notifications are sent to users in a single email in conjunction with each of the three yearly MetaCyc releases.
Users can define areas of interest in several ways:
By entering one or more specific pathways of interest
By defining a SmartTable listing pathways of interest
By entering a pathway class of interest.
For example, after specifying the MetaCyc pathway class ‘Sulfur Compounds Metabolism’, users will receive updates about new or revised pathways that are classified under that class. To enter new update-notification requests, users log into their BioCyc account, navigate to the desired pathway, pathway class, or SmartTable page, and click the ‘Get Email Notifications of Updates’ command in the right-sidebar Operations menu.
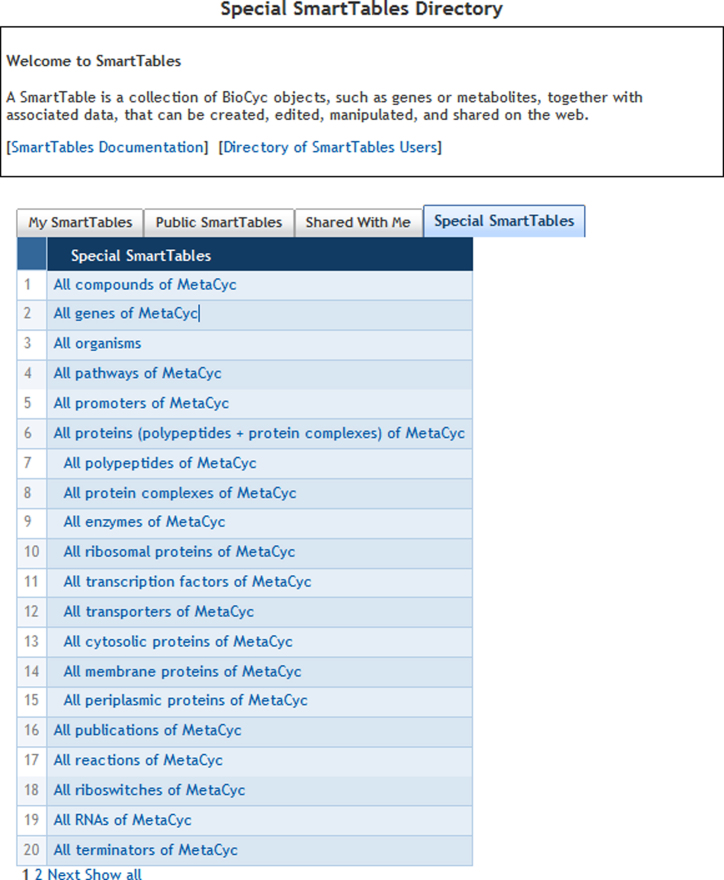
SmartTables
SmartTables provide a powerful way for users to arrange and manipulate data in MetaCyc and other PGDBs. Although SmartTables are not a new feature in MetaCyc, we would like to mention them to ensure that all users are familiar with this powerful tool. SmartTables are spreadsheet-like structures that can contain both PGDB objects and other data such as numbers or text. Like a spreadsheet, a SmartTable is organized by rows and columns that users can add to or delete. A typical SmartTable contains a set of PGDB objects in the first column (e.g. a set of compounds generated by a search). The other columns contain properties of the object (e.g., the chemical composition of the compounds) or the result of a transformation (e.g. the reactions in which these compounds participate).
While users can create their own SmartTables, several SmartTables are already available for users, including such tables as all compounds in MetaCyc, all pathways in MetaCyc, all polypeptides of MetaCyc, etc. (see Figure 3). You will find these special tables under the SmartTable menu (SmartTables → Special SmartTables).
Figure 3.
MetaCyc contains a number of pre-formed SmartTables that provide access to results of popular searches.
Protein sequence data
Previously, one difference between the proteins curated in MetaCyc and those in organism-specific PGDBs was that MetaCyc proteins did not contain sequence information, preventing users from performing BLAST searches within MetaCyc. As of 2017, sequence data is available for all MetaCyc proteins that have links to the UniProt database (15), which in version 21.1 of MetaCyc, comprised 10,560 proteins (∼79% of all MetaCyc polypeptides). When browsing such a protein, users can now select the command ‘Show Sequence at UniProt’ from the Operations menu to display the sequence in FASTA format. In addition, BLAST searches have been enabled in MetaCyc, which is done by selecting the BLAST search command from the Search menu. The results of the BLAST search are provided in an html document that provides links to the MetaCyc pages of the candidate proteins, enabling users to quickly navigate their way from a protein sequence to pages describing reactions and pathways associated with related proteins.
Search for reactions by substrates
This command, which enables users to search for reactions by specifying one or more substrates, has been expanded to enable specifying on which side of the reaction different substrates appear (relative to each other). This type of search, which to the best of our knowledge is unavailable elsewhere, enables users to specify more complex search parameters. For example, searching separately for dechlorination reactions that utilize water (water on one side, chlorine on the other side) or dechlorination reactions that produce water (water and chlorine on the same side) is now possible.
Set MetaCyc as the default database
Users who employ MetaCyc most of the time (as opposed to other PGDBs) can now have MetaCyc automatically selected whenever they log into the BioCyc website. To do so, select My Account from the top right corner, click the ‘Database Selection’ tab, and then choose MetaCyc.
SUBSCRIPTION MODEL FOR BIOCYC ACCESS
In our previous papers in the database issue of Nucleic Acids Research, we described the MetaCyc database together with the BioCyc PGDB collection (5). As of 2017 SRI International has adopted a subscription-based model for BioCyc access. MetaCyc, as well as the EcoCyc PGDB [the PGDB for Escherichia coli K-12, (16)], remain freely available to all and do not require a subscription.
Because of ongoing difficulties in securing government funds for database curation, we moved to a subscription-based model in the hope of generating funds that would permit us to curate high-quality databases for more organisms, such as important pathogens, biotechnology workhorses, model organisms and promising hosts for biofuels development. More information about the subscription model is available at http://www.phoenixbioinformatics.org/biocyc/index.html.
HOW TO LEARN MORE ABOUT METACYC AND BIOCYC
The MetaCyc.org website provides several informational resources, including an online guide for MetaCyc (http://www.metacyc.org/MetaCycUserGuide.shtml); a guide to the concepts and science behind the Pathway/Genome Databases (http://biocyc.org/PGDBConceptsGuide.shtml); and instructional webinar videos that describe the usage of MetaCyc, BioCyc and Pathway Tools (http://biocyc.org/webinar.shtml). We routinely host workshops and tutorials (on site and at conferences) that provide training and in-depth discussion of our software for both beginning and advanced users. To stay informed about the most recent changes and enhancements to our software, please join the BioCyc mailing list at https://biocyc.org/subscribe.shtml. A list of our publications is available online at https://biocyc.org/publications.shtml.
DATABASE AVAILABILITY
The MetaCyc database is freely and openly available to all. See https://biocyc.org/download.shtml for download information. New versions of the downloadable data files and the MetaCyc website are released three times per year. Access to the website is free; users are required to register for a free account after viewing more than 30 pages in a given month.
ACKNOWLEDGEMENTS
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
FUNDING
National Institute of General Medical Sciences of the National Institutes of Health (NIH) [GM080746, GM077678, GM75742]. Funding for open access charge: National Institute of General Medical Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016; 44:D471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karp P.D., Paley S.M., Krummenacker M., Latendresse M., Dale J.M., Lee T.J., Kaipa P., Gilham F., Spaulding A., Popescu L. et al. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief. Bioinform. 2010; 11:40–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karp P.D., Latendresse M., Paley S.M., Krummenacker M., Ong Q.D., Billington R., Kothari A., Weaver D., Lee T., Subhraveti P. et al. Pathway Tools version 19.0 update: software for pathway/genome informatics and systems biology. Brief. Bioinform. 2016; 17:877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karp P.D., Latendresse M., Caspi R.. The pathway tools pathway prediction algorithm. Standards Genomic Sci. 2011; 5:424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karp P.D., Billington R., Caspi R., Fulcher C.A., Latendresse M., Kothari A., Keseler I.M., Krummenacker M., Midford P.E., Ong Q. et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinformatics. 2017; doi:10.1093/bib/bbx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vallenet D., Calteau A., Cruveiller S., Gachet M., Lajus A., Josso A., Mercier J., Renaux A., Rollin J., Rouy Z. et al. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 2017; 45:D517–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mazourek M., Pujar A., Borovsky Y., Paran I., Mueller L., Jahn M.M.. A dynamic interface for capsaicinoid systems biology. Plant Physiol. 2009; 150:1806–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schlapfer P., Zhang P., Wang C., Kim T., Banf M., Chae L., Dreher K., Chavali A.K., Nilo-Poyanco R., Bernard T. et al. Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 2017; 173:2041–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walsh J.R., Schaeffer M.L., Zhang P., Rhee S.Y., Dickerson J.A., Sen T.Z.. The quality of metabolic pathway resources depends on initial enzymatic function assignments: a case for maize. BMC Syst. Biol. 2016; 10:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Evsikov A.V., Dolan M.E., Genrich M.P., Patek E., Bult C.J.. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009; 10:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jankowski M.D., Henry C.S., Broadbelt L.J., Hatzimanikatis V.. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys. J. 2008; 95:1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alberty R.A. Thermodynamics of Biochemical Reactions. 2003; Wiley InterScience. [Google Scholar]
- 13. McDonald A.G., Tipton K.F.. Fifty-five years of enzyme classification: advances and difficulties. FEBS J. 2014; 281:583–592. [DOI] [PubMed] [Google Scholar]
- 14. Green M.L., Karp P.D.. Genome annotation errors in pathway databases due to semantic ambiguity in partial EC numbers. Nucleic Acids Res. 2005; 33:4035–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keseler I.M., Mackie A., Santos-Zavaleta A., Billington R., Bonavides-Martinez C., Caspi R., Fulcher C., Gama-Castro S., Kothari A., Krummenacker M. et al. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017; 45:D543–D550. [DOI] [PMC free article] [PubMed] [Google Scholar]