Abstract
Here we present an updated version of the AU-Rich Element Database (ARED-Plus) that is freely available at http://brp.kfshrc.edu.sa/ared. AREs are conserved sequence elements that were first discovered in the 3′UTR of mammalian transcripts. Over the past years, we compiled a series of ARE databases that revealed the extent and wide distribution of ARE-containing genes. For this update, we adopted an optimized search algorithm with improved specificity and sensitivity in ARE selection. The designation of the different ARE clusters was simplified by directly correlating the number of the ARE cluster to the number of overlapping AUUUA pentamers. Additionally, the new database was expanded to include genes with intronic AREs (pre-mRNAs) and their characteristics since recent observations reported their abundance and biological significance. Several enhancements were incorporated such as customized column view, additional search options and live search functionalities. The new version includes links to AREsite and AREScore, two related ARE assessment algorithms for further evaluation of the ARE characteristics. ARED-Plus now contains an updated repertoire of AREs in the human transcriptome that may be useful in several research fields.
INTRODUCTION
Post-transcriptional regulation is an important step in gene expression, and contributes to the rapid and transient nature of many biological responses. Essential for these control checkpoints are AU-rich elements (AREs) which are cis-acting elements that were initially found in the 3′-untranslated regions (3′UTR) of the mRNA and regulate its decay and translation. They are bound by several RNA-binding proteins such as tristetraprolin (TTP/ZFP36), AUF1 (HNRPD), and HuR (ELAVL1), KSRPH among others (1–3). Using a computational approach, we previously expanded the number of ARE-containing transcripts to nearly 8% of human mRNAs and the ARE-database (ARED) was established (4). Improved versions of the database were compiled that benefited from advancements in available data sources and included functional classification and gene ontology (ARED 2.0 and ARED 3.0). Later work expanded the database to the mouse and rat mRNAs and showed significant levels of ARE conservation across orthologous genes, albeit with some notable exceptions (ARED organism) (5).
There are recent advances in the field including the association of ARE-binding proteins such as AUF1, HuR, TTP and ZFP36L1 with ARE-like or U-rich sequence elements that are located in the introns of human genes (6–11). We also found that AREs are abundant in introns and the expression of intronic-ARE or U-rich coding genes can be affected by HuR (6–8, and our manuscript in revision). Therefore, we compiled this new update that not only represents updated information on ARE-mRNAs and also pre-mRNAs, but features an improved search algorithm and other web and link enhancements. The database now comprises detailed information of the 22% of human 3′UTRs as ARE-mRNAs and 25% of human introns (ARE pre-mRNAs) that correspond to more than half of the human genes.
IMPROVED SEARCH ALGORITHM AND SIMPLIFIED CLUSTER DESIGNATION
We adopted a new algorithm with improved sensitivity and specificity for extracting the ARE-containing transcripts. First, unlike the previous ARED versions, the designation of the ARE clusters is now directly correlated with the number of overlapping AUUUA pentamers (Clusters, Clusters I to V, Table 1). A typical ARE contains a least one core AUUUA pentamer in an AU-rich context. The 9-mer UUAUUUAUU and the 13-mer WWUUAUUUAUUWW patterns have been suggested as the minimal requirement for functionality (8–10). Overlapping reiterations of the core pentamer potentiate the activity of the element. Several different ARE search algorithms were investigated that allow the extraction of Cluster 1 AREs which contains the single AUUUA pentamer in an AU-rich surrounding: W1W2W3W4(A5U6U7U8A9)W10W11W12W13 (W = A or U; Supplementary Table S1). Sensitivity was assessed against a reference list of 93 experimentally established AREs, (Supplementary Table S2). Specificity for the 3′UTR and intronic regions was assessed against the 5′UTR and coding regions of genes. We explored eight conditions with varying levels of stringency to search for the AREs of cluster 1 (Supplementary Table S1). The following conditions proved optimal and were selected: no mismatch in the core region (AUUUA), two or more of the W at positions 3, 4, 10 and 11 are restricted to Us and up to two mismatches are allowed at positions 1, 2, 12 and 13 and at least one of positions of 4 or 10 must be U. These conditions showed highest sensitivity against an ARE-mRNA reference list (matched 98.3% of the ARE reference list), and high specificities (absent from 96.55% of coding regions, Supplementary Table S1). Other clusters are shown in Table 1.
Table 1. Patterns and conditions of ARE clusters.
| CLUSTER | Pattern | Conditions |
|---|---|---|
| 1 | W1W2W3W4(A5U6U7U8A9)W10W11W12W13 | • 2 or more of positions 3, 4, 10, 11 restricted to Us |
| • Only one mismatch allowed at positions 1, 2, 12,13 | ||
| • At least One position 4 or 10 is U | ||
| 2 | W(TATTTATTTA)WW or WW(ATTTATTTAT)W | • Derived from cluster 1 |
| • Two mismatches allowed outside the core | ||
| 3 | ATTTATTTATTTA | No mismatch is allowed |
| 4 | ATTTATTTATTTATTTA | No mismatch is allowed |
| 5+ | +ATTTATTTATTTATTTATTTA+ | One mismatch is allowed |
The number of the ARE cluster directly correlates to the number of overlapping AUUUA pentamer.
The new algorithm was used to compile ARE-mRNAs (3′UTR-centric) and ARE pre-mRNA (intron-centric) in the new ARED (depicted as ARED-Plus). According to this latest screen, the ARED Plus database contains 4884 mRNAs (ARE-mRNAs) (22% of the human genes) and 12 822 pre-mRNAs (ARE-pre-mRNAs) which corresponds to 58% of the human genes.
WEBSITE DESCRIPTION
The new website retains all the features of the previous versions with notable additions and improvements. Most notably, it allows the user to search for AREs in the introns in addition to the 3′UTR regions. The investigator has the option to select an ARE cluster or several clusters which correlate with the number of overlapping pentamers from each category. An auto-complete feature facilitates the submission of an indefinite number of keywords. Three options are available for a search query: Simple, Advanced and List. The simple option has one field for keywords, the advanced option allows specified options/categories to place additional keywords, and the list search allows the user to insert a list of identifiers. The output table has customized column visibility and can be either uploaded in CSV, PDF and Excel formats or printed. The table contains a real time search function that allows the user to retain the rows that contain the search item.
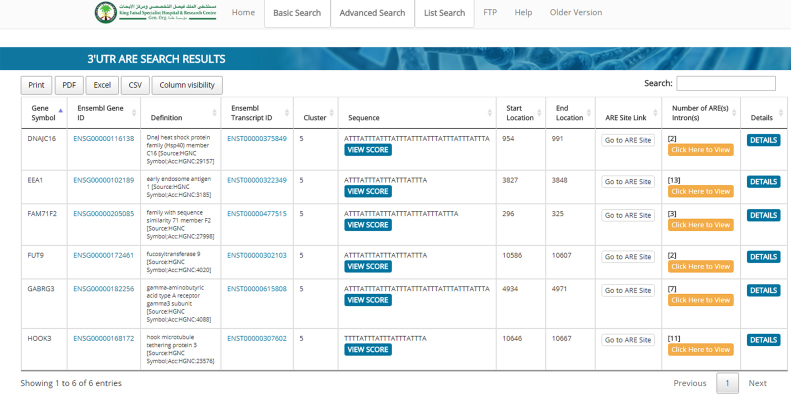
The website allows the user to select for either the 3′UTR AREs, intron AREs or both. The features displayed for a 3′UTR search in the table include the following: Gene Symbol, Ensembl Gene ID, definition, Ensembl Transcript ID, ARE Cluster, ARE Sequence, ARE Start and end locations. For further sequence details of ARE motifs, a link to the AREsite 2.0 was created (http://rna.tbi.univie.ac.at/AREsite2/welcome; (12). The ARED-Plus website has also an additional ARE evaluation feature by linkage to ARE score website (http://arescore.dkfz.de/arescore.pl) which provides numerical assessment of the ARE potency (13). Each gene record has an additional detail link to additional information such as GO annotations, RefSeq, UNIGENE and Entrez IDs (Example is shown in Figure 1).
Figure 1.
Output table of a basic 3′UTR and intron ARE search. Typically, the output table contains the gene names, Ensembl gene IDs, genes definition, Ensembl transcript IDs, ARE-clusters and sequence, in the sequence pane a link to ARE SCORE is available, start and end location of the ARE in the transcript, a link to the ARE site and a details link to additional information such as GO annotations, RefSeq, UNIGENE and Entrez IDs. If the gene contains intronic AREs their number will be displayed along with a link that directs the user to them.
Another feature of ARED-Plus is the inclusion of ARE introns. For an intronic ARE display, additional features include the chromosome number, the locations of the ARE and the intron. If a gene contains more than one intron with AREs, the output will show this gene in multiple displays for each ARE. If both 3′UTR AREs, intron AREs are selected the output row in the table will contain the features mentioned above for 3′UTR ARE with an additional column entitled ‘Number of ARE-Introns’. This column displays the number of intronic AREs present for a specific gene and contains a link that directs the user to the intronic AREs and their features. A detailed instruction file is available that guides users.
CONCLUSIONS AND FINAL REMARKS
The new version of the ARED database was developed with updated datasets for AU-rich mRNAs (3′UTR-centric) and inclusion of ARE introns with enhanced search strategy and web features. ARED-plus now includes more than 63% of human genes compared to only 11% of previous versions that comprised only AREs of the 3′UTR. It should be noted that the classical function of AREs, i.e. default mRNA decay, translational inhibition, and response to both destabilizing and stabilizing promoting RNA-binding proteins happen largely with ARE-mRNAs rather than pre-ARE-mRNAs. However, ARE pre-mRNAs are largely responsive to the stability promoting RBP HuR and probably to other RNA-binding proteins in absence of other functions (6,7, and manuscript in revision). Other improvements include a new search algorithm with improved sensitivity and specificity in ARE selection as well as an upgraded user platform that facilitates operation. This update accounts for the progress of the last years and allows global research community to explore the human genome for AREs in a comprehensive display. Deviations in the intricate regulation of ARE-mediated processes have been linked to conditions such as cancer and chronic inflammatory diseases (14). Thus, ARED-Plus database should prove useful to the biomedical research community across different fields.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
King Faisal Specialist Hospital and Research Centre [RAC # 2110012]. Funding for open access charge: King Faisal Specialist Hospital and Research Centre.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dean J.L., Sully G., Clark A.R., Saklatvala J.. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell. Signal. 2004; 16:1113–1121. [DOI] [PubMed] [Google Scholar]
- 2. von Roretz C., Di Marco S., Mazroui R., Gallouzi I.E.. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley interdiscipl. Rev. RNA. 2011; 2:336–347. [DOI] [PubMed] [Google Scholar]
- 3. Schott J., Reitter S., Philipp J., Haneke K., Schafer H., Stoecklin G.. Translational regulation of specific mRNAs controls feedback inhibition and survival during macrophage activation. PLoS Genet. 2014; 10:e1004368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bakheet T., Frevel M., Williams B.R., Greer W., Khabar K.S.. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001; 29:246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Halees A.S., El-Badrawi R., Khabar K.S.. ARED Organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2008; 36:D137–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lebedeva S., Jens M., Theil K., Schwanhausser B., Selbach M., Landthaler M., Rajewsky N.. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011; 43:340–352. [DOI] [PubMed] [Google Scholar]
- 7. Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M. Jr, Tuschl T., Ohler U., Keene J.D.. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011; 43:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sedlyarov V., Fallmann J., Ebner F., Huemer J., Sneezum L., Ivin M., Kreiner K., Tanzer A., Vogl C., Hofacker I. et al. Tristetraprolin binding site atlas in the macrophage transcriptome reveals a switch for inflammation resolution. Mol. Syst. Biol. 2016; 12:868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoon J.H., De S., Srikantan S., Abdelmohsen K., Grammatikakis I., Kim J., Kim K.M., Noh J.H., White E.J., Martindale J.L. et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 2014; 5:5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Newman R., Ahlfors H., Saveliev A., Galloway A., Hodson D.J., Williams R., Besra G.S., Cook C.N., Cunningham A.F., Bell S.E. et al. Maintenance of the marginal-zone B cell compartment specifically requires the RNA-binding protein ZFP36L1. Nat. Immunol. 2017; 18:683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mukherjee N., Jacobs N.C., Hafner M., Kennington E.A., Nusbaum J.D., Tuschl T., Blackshear P.J., Ohler U.. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 2014; 15:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fallmann J., Sedlyarov V., Tanzer A., Kovarik P., Hofacker I.L.. AREsite2: an enhanced database for the comprehensive investigation of AU/GU/U-rich elements. Nucleic Acids Res. 2016; 44:D90–D95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spasic M., Friedel C.C., Schott J., Kreth J., Leppek K., Hofmann S., Ozgur S., Stoecklin G.. Genome-wide assessment of AU-rich elements by the AREScore algorithm. PLoS Genet. 2012; 8:e1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khabar K.S. Hallmarks of cancer and AU-rich elements. Wiley Interdiscipl. Rev. RNA. 2017; 8, doi:10.1002/wrna.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.