Abstract
AutDB is a deeply annotated resource for exploring the impact of genetic variations associated with autism spectrum disorders (ASD). First released in 2007, AutDB has evolved into a multi-modular resource of diverse types of genetic and functional evidence related to ASD. Current modules include: Human Gene, which annotates all ASD-linked genes and their variants; Animal Model, which catalogs behavioral, anatomical and physiological data from rodent models of ASD; Protein Interaction (PIN), which builds interactomes from direct relationships of protein products of ASD genes; and Copy Number Variant (CNV), which catalogs deletions and duplications of chromosomal loci identified in ASD. A multilevel data-integration strategy is utilized to connect the ASD genes to the components of the other modules. All information in this resource is manually curated by expert scientists from primary scientific publications and is referenced to source articles. AutDB is actively maintained with a rigorous quarterly data release schedule. As of June 2017, AutDB contains detailed annotations for 910 genes, 2197 CNV loci, 1060 rodent models and 38 296 PINs. With its widespread use by the research community, AutDB serves as a reference resource for analysis of large datasets, accelerating ASD research and potentially leading to targeted drug treatments. AutDB is available at http://autism.mindspec.org/autdb/Welcome.do.
INTRODUCTION
Autism Spectrum Disorders (ASD) are characterized by symptoms in two behavioral domains: social communication and interaction, and restricted interests and repetitive behaviors (1). However, the phenotypic profile of ASD exceeds far beyond the core behavioral domains and includes diverse medical co-morbidities (2). The clinical complexity of ASD is mirrored at the level of genetic heterogeneity. Hundreds of genes and chromosomal loci are known to be associated with the disorder. To facilitate visualization and analysis of the vast genetic heterogeneity underlying ASD, we created AutDB, the Autism Gene Database, in 2007 as a publicly available, manually curated, online resource for genes linked to ASD (3). AutDB is licensed to the Simons Foundation as SFARI Gene (4,5).
In recent years, several large-scale, collaborative research efforts have focused on characterizing the genetic risk architecture of ASD in well-defined cohorts using genome-wide methodologies (6,7). These studies have identified several ASD risk genes and copy number variants (CNV) with rigorous statistical support (8). A growing list of promising candidate genes for ASD is regularly reported in the scientific literature in an attempt to define the genetic underpinnings of ASD (9,10). The advances in ASD genetics have led researchers to investigate the biological role of the ASD risk genes by generating appropriate animal models or defining relevant biomolecular interaction patterns. AutDB has closely followed this trend in autism research by incorporating new datasets in the form of modules that are integrated with the ASD gene entries to facilitate multimodal exploration. In its current form, four interconnected modules comprise AutDB: (i) Human Gene; (ii) CNV; (iii) Animal Model; and (iv) Protein Interaction (PIN). To provide an exhaustive coverage of autism research, AutDB curates information from both high- and low-throughput studies into a standardized resource. Together, AutDB has grown rapidly in content over the last 10 years with a regular quarterly release schedule providing researchers immediate access to new data shortly after publication.
DATABASE DESIGN AND CONTENT
Modular architecture of AutDB
The inaugural release of AutDB included only the Human Gene module, which incorporated all known genes linked to ASD found through genetic association or mutational analysis (3). The Human Gene module was the first database to include genes containing both rare and common variants associated with a complex disorder. However, to understand the biological impact of ASD risk genes, we have updated and expanded AutDB with a systems biology approach. Through a primary focus on ASD risk genes, we have added new datasets with information reported on CNVs, animal models and PINs into an integrated, multi-modular framework (Figure 1). An overview of the data in individual modules is shown in Table 1.
Figure 1.
Modular architecture of AutDB. The Human Gene module is the central component of AutDB collecting genetic risk factors for ASD including rare and common variants identified from high- as well as low-throughput studies. The CNV module includes a comprehensive collection of deletions and duplications of chromosomal loci identified in ASD individuals. To comprehend the functional significance of genetic risk factors, two additional sources of data are included: animal models and protein interactomes of ASD-associated genes. Notably, the diverse structures of genetic and functional data are captured and integrated using annotation models that preserve the biological meaning of the entities. Module-specific front pages provide easy ways for navigation across the other modules in AutDB.
Table 1. Overview of data content in AutDB (June 2017).
| Module | Entry | Count | |
|---|---|---|---|
| Human Gene | Genes | 910 | |
| Rare variants | 8939 | ||
| Common variants | 1203 | ||
| Curated references | 1664 | ||
| Additional references | 1413 | ||
| Animal Model | Genetic models | Mouse | 1012 |
| Rat | 23 | ||
| Inducers | 71 | ||
| Induced models | Mouse | 109 | |
| Rat | 186 | ||
| Curated references | Mouse | 600 | |
| Rat | 131 | ||
| CNV | CNV loci | 2197 | |
| Curated references | 531 | ||
| PIN | Interactions | 38 296 | |
| Curated references | 2694 |
Human Gene module
The Human Gene module, cataloging genes associated with ASD, has undergone major enhancements since its initial release. The original framework of AutDB included curation of genes based on evidence from genetic association studies, rare single gene mutations, syndromic autism and genes with functional support (Basu, 2009). In the succeeding years, this annotation model has been expanded to include detailed information pertaining to rare and common genetic variants associated with ASD. Currently, AutDB includes a broad range of potentially pathogenic variants such as single nucleotide variants (SNVs), insertion-deletion variants (indels) and structural variations disrupting a single gene. Rare variant-specific information displayed in the Human Gene module includes Variant Type, Inheritance Pattern, Inheritance Association and Family Type. Common variant-specific information displayed in the Human Gene module includes Polymorphism, SNP ID, Population origin and Population Stage. We recently developed a gene ranking algorithm utilizing the cumulative strength of evidence for variants identified in ASD-associated genes (11). Here, we incorporate this work into our annotation model to show the ranking of ASD genes for the first time within the Human Gene module.
Animal Model module
Model animals are vital to study the contribution that discrete genetic differences have towards the development of complex phenotypes related to human disorders. The Animal Model module was introduced in AutDB to systematically record anatomical, behavioral and physiological data from mouse models arising from ASD-linked genes (12). In the past few years, however, the field has witnessed an exponential rise of gene-targeted ASD models in terms of the complexity of the models, the number of articles and their publication in high-impact journals. Consequently, this module has undergone significant expansion with respect to curation of new model types. At present, AutDB includes both mouse and rat models of ASD. All models generated from the manipulation of ASD genes are fully integrated with the corresponding entry in the Human Gene module.
A novel feature of the Animal Model module is the inclusion of a dataset encompassing environmentally induced models of ASD. We made this development in response to the increasing scientific evidence for a multifactorial etiology for autism (13). A landmark study in 2014 convincingly indicated that variance in autism liability has contributions generated from both genetic and non-genetic factors (14). Although the contribution of non-genetic risk factors toward ASD is poorly understood, a long list of environmental agents has been linked to ASD through epidemiological studies. Researchers are generating an increasing number of induced models to elucidate the role of non-genetic factors in ASD (15,16). In the AutDB annotation model, the environmental ‘agent’ used to induce phenotypic or behavioral parallels of ASD in wild type animals is classified as ‘Chemical’ or ‘Biological’. Notable examples of non-genetic ASD risk factors successfully modeled in rodents that are curated in AutDB include advanced paternal age (17), maternal immune activation (18), and exposure to viruses (19) or drugs during fetal development (20).
Continuing to follow cutting edge research trends in ASD, we began to annotate data from ‘rescue’ animal models where pharmaceutical, procedural or genetic interventions restore a phenotype of ASD models. Rescue animal models include animals treated with protein factors, like insulin like growth factor-1 (IGF-1), that alleviate abnormal behavioral and neurophysiological phenotypes. The rescue models add another dimension to the Animal Model module, forming a bridge to translational clinical research.
Protein Interaction (PIN) module
The PIN module of AutDB is a comprehensive, up-to-date reference for all known PINs of gene products associated with ASD. Unlike other AutDB modules, PIN data queries are not limited to the field of ASD research. Instead, we probe the general biological literature to identify and curate PINs for ASD risk genes, with a particular emphasis on neuron-specific interactions. The PIN annotation model specifies three types of binding (protein binding, RNA binding and promoter binding) and three types of regulation (protein modification, direct regulation and autoregulation). Relationships can be activating, inhibiting or neutral. Each interaction is linked to the source article and annotated based on a detailed standardized meta-data on experimental paradigms, cell type specificity, species specificity and UNIPROT IDs of all interacting proteins. Details of protein manipulations reported in the studies, such as isoform specificities and fusion proteins are also included in the annotation of each PIN entry. Finally, interaction data in PIN are fully integrated with entries in the Human Gene module.
Copy number variant (CNV) module
The CNV module of AutDB represents a comprehensive collection of all known CNVs associated with ASD. The CNV data are organized based on the chromosomal loci in which they were observed in the annotated report. For any given CNV locus, a published report can be characterized as either ‘major’ or ‘minor’. A designation of ‘major’ indicates that an independent secondary methodology was used to confirm or validate at least one CNV within the locus of interest following its initial discovery. On the other hand, if the report-class for a CNV locus is listed as minor, no subsequent validation or confirmation was performed following the initial discovery of any CNV at the locus of interest.
Data for a given CNV locus are presented in a table under four tabs: CNV summary, population data, individual data and animal model. The CNV summary page includes a summary of the evidence linking CNVs at a particular locus to ASD or a related neurodevelopmental/neuropsychiatric disorder. Additional locus information is based on external links to the corresponding CNV summary pages of two commonly utilized genome browsers (the UCSC and NCBI Genome Browsers) and the DECIPHER database (http://decipher.sanger.ac.uk). The CNV module organizes data based both at the cohort and individual levels. Cohort-level data include cohort size, age, gender and geographical ancestry, as well as the CNV discovery and validation methodology used in each study. Individual-level data include detailed clinical and cognitive profiles, as well as information on CNV inheritance, segregation of the CNV with disease and the RefSeq gene content of each CNV. Finally, the animal model page provides detailed phenotypic information on mouse models engineered to model CNVs identified in ASD individuals.
DATA CURATION, MAINTENANCE AND RELEASE
AutDB strives to build the most comprehensive collection of ASD-associated genes, CNVs, animal models and PINs extracted from current scientific publications. The datasets are systematically built following established guidelines for the curation process. Scientist annotators continuously search, analyze and synthesize ASD-related information from published scientific literature. Developed at MindSpec, the AutDB annotation model is deeply rooted in the biology of ASD, requiring integration of diverse types of knowledge. The entire content of AutDB is manually annotated from scientific reports.
Identification of relevant scientific literature
Our strategy involves exhaustive searches for relevant journal articles by automated, timed PubMed queries to build a comprehensive collection. The automated searches are supplemented by annotator-initiated regular searches of high-impact journal websites for pre-publication release of ‘in press’ articles. The collected papers are first analyzed for their data content and selected on the basis of established inclusion/exclusion criteria. Overall, a substantial proportion of papers gathered by automated searches are rejected due to insufficient data. Finally, the selected papers are critically assessed based on module-specific annotation protocols.
Annotation process
The full text of each selected paper is extensively reviewed including tables, figures and supplementary material embedded within the publication for data extraction. Using a multilevel annotation protocol, information from the article is extracted using the standardized AutDB-scientific-annotation model that is specific for each module. New AutDB entries are built by integrating information from all references that are used during the curation procedure. For existing entries, new information extracted from a recent reference is added to update the entry.
All completed entries are passed through a series of automated checks that verify a number of annotation rules. Any detected errors are sent back to the annotator and corrected. Next, a manual review is undertaken to ensure that all relevant information and citations in the input file are valid. These data are then merged with the existing curated datasets and saved as a master file. During the merging process, our software validates each entry using an extensive set of rules along with referential integrity of primary keys performing the process of de-duplication. Unit testing is enforced for any data or code changes.
Database releases
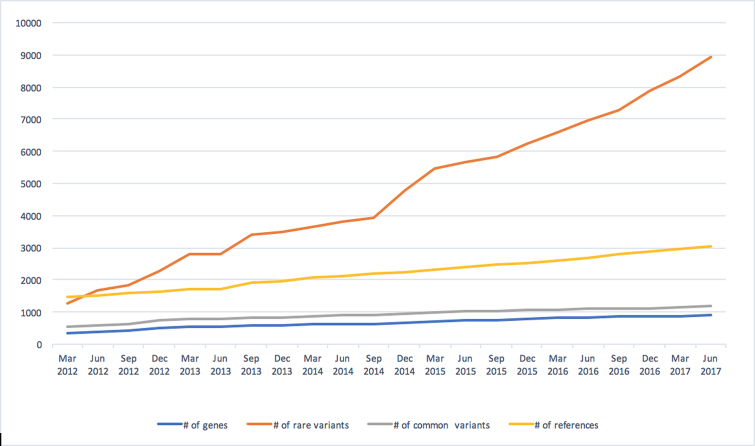
AutDB is rigorously maintained by systematic updates of each modules and quarterly database releases. As shown in Figure 2, data content within Human Gene module of AutDB has grown rapidly keeping pace with the growth of autism research. For example, from March (Q1) 2012 to June (Q2) 2017, the number of genes in the Human Gene module nearly tripled so that 910 genes were linked to ASD as of 30 June 2017 (Figure 2). While the number of rare and common genetic variants identified in ASD-associated genes both increased since the first quarter of 2012, the more dramatic rise in the number of rare variants compared to common variants over that length of time reflects the growing use of high-throughput genetic screening technologies in analyzing ASD cohorts.
Figure 2.
Growth of Human Gene module of AutDB (March 2012–June 2017). The number of genes associated with ASD increased from 328 to 910 with concomitant increase of rare and common variants from 1282 to 8938 and 549 to 1203, respectively. Rare variants are defined as those with a population frequency <1%; common variants are defined as those found in the general population at a frequency of ≥1%.
DATA ACCESS
AutDB is a custom-built resource that be can be easily accessed through the public website http://autism.mindspec.org/autdb/Welcome.do. The genetic and functional data in AutDB are presented through a user-friendly interface design that allows easy exploration of the curated data. Within a tab-based structure, users can easily navigate between genes and their associated CNVs, corresponding animal models and protein interactomes, gaining a current and composite snapshot of interdisciplinary research on ASD. The autonomy of individual modules is fully retained in AutDB; users can interrogate, create subsets and download single source, module-specific dataset for further analysis. Through the Advanced Search page, users can select a specific dataset (Human Gene, CNV, Animal Model or PIN) and query the database with standardized gene symbols (e.g. MECP2), common gene names (oxytocin), gene category (e.g. rare, syndromic, association, functional, other), chromosome number (e.g. 1–22, X, Y), chromosome band (e.g. 16p11.2). Moreover, the homepage includes a link to the MindSpec Reading Room that is periodically updated with popular articles on basic ASD-relevant biology and concise summaries of current findings in ASD research.
AutDB AT 10 YEAR MARK
AutDB was created to serve as a reference resource to help unravel the biological underpinnings of ASD. Since its debut in 2007, AutDB has been used extensively by the ASD research community as indicated by the rapid growth of its citations over the past years. Importantly, the curated datasets of AutDB are used for establishing new roles of ASD risk genes or deciphering large-scale datasets.
At present, fast-paced advances in genomic technology and their application on large cohorts are leading to the enhanced understanding of ASD biology. Consequently, our scientific annotation model requires continuous assessments and updates based on newly reported data. A direct outcome of the updated annotation model is a requirement for the harmonization of previously annotated data. Given the lack of uniform standards of data reporting in scientific publications, we continue to standardize literature-extracted data using controlled vocabulary established by bioinformatics consortiums. Database maintenance with strict quarterly releases is a critical component of this phase.
Although several new features have been introduced into the updated version of AutDB to incorporate current research on autism and to enhance the accessibility of the curated data with the user in mind, our team is also focused on developing tools to enhance the utility the database. To provide an assessment framework to our users, we have recently developed a gene ranking algorithm based on the cumulative strength of evidence for each gene cataloged in AutDB (11). Our methodology is centered on systematic evaluation of variants implicated in ASD taking into consideration the significance of genetic association, family structure and inheritance pattern (de novo or transmitted) and type (loss-of-function, missense, deletion or duplications in a gene). One limitation of our gene-based approach is that it excludes large, multigenic CNVs associated with ASD. In a future endeavor, our goal is to introduce new criteria for assessment of CNVs curated in AutDB.
CONCLUDING REMARKS
To sum up, the exponential growth of diverse research modalities that is pushing the boundaries of our understanding of ASD and related biology requires thoughtful open-minded consideration and continuous integration. It is our mission at MindSpec, to continue to utilize detailed scientific curation to provide an accessible and exhaustive resource for bioinformatics analyses, basic and translational scientific research, and to raise public awareness. This approach is targeted at accelerating the pace of development in effective behavioral and chemotherapeutic agents that will help manage, if not prevent or cure, the complex afflictions of ASD, in the foreseeable future.
ACKNOWLEDGEMENTS
We thank the Simons Foundation for their generous support. We would also like to acknowledge Alan Packer of the Simons Foundation Autism Research Initiative for helpful feedback and support for this project.
FUNDING
AutDB is funded by the Simons Foundation, which licenses it as SFARI Gene http://gene.sfari.org/. Funding for open access charge: MindSpec Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1. American Psychiatric Association American Psychiatric Association. DSM-5 Task Force. Diagnostic and Statistical Manual of Mental Disorders: DSM-5. 2013; xliv:5th ednWashington, D.C.: American Psychiatric Association; 947. [Google Scholar]
- 2. Doshi-Velez F., Ge Y., Kohane I.. Comorbidity clusters in autism spectrum disorders: an electronic health record time-series analysis. Pediatrics. 2014; 133:e54–e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basu S.N., Kollu R., Banerjee-Basu S.. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009; 37:D832–D836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abrahams B.S., Arking D.E., Campbell D.B., Mefford H.C., Morrow E.M., Weiss L.A., Menashe I., Wadkins T., Banerjee-Basu S., Packer A.. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism. 2013; 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banerjee-Basu S., Packer A.. SFARI Gene: an evolving database for the autism research community. Dis. Model. Mech. 2010; 3:133–135. [DOI] [PubMed] [Google Scholar]
- 6. Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012; 485:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iossifov I., Ronemus M., Levy D., Wang Z., Hakker I., Rosenbaum J., Yamrom B., Lee Y.H., Narzisi G., Leotta A. et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012; 74:285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanders S.J., He X., Willsey A.J., Ercan-Sencicek A.G., Samocha K.E., Cicek A.E., Murtha M.T., Bal V.H., Bishop S.L., Dong S. et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015; 87:1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stessman H.A., Xiong B., Coe B.P., Wang T., Hoekzema K., Fenckova M., Kvarnung M., Gerdts J., Trinh S., Cosemans N. et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 2017; 49:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. RK C.Y., Merico D., Bookman M., J L.H., Thiruvahindrapuram B., Patel R.V., Whitney J., Deflaux N., Bingham J., Wang Z. et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017; 20:602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larsen E., Menashe I., Ziats M.N., Pereanu W., Packer A., Banerjee-Basu S.. A systematic variant annotation approach for ranking genes associated with autism spectrum disorders. Mol. Autism. 2016; 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kumar A., Wadhawan R., Swanwick C.C., Kollu R., Basu S.N., Banerjee-Basu S.. Animal model integration to AutDB, a genetic database for autism. BMC Med. Genomics. 2011; 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hallmayer J., Cleveland S., Torres A., Phillips J., Cohen B., Torigoe T., Miller J., Fedele A., Collins J., Smith K. et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry. 2011; 68:1095–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaugler T., Klei L., Sanders S.J., Bodea C.A., Goldberg A.P., Lee A.B., Mahajan M., Manaa D., Pawitan Y., Reichert J. et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014; 46:881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ingram J.L., Peckham S.M., Tisdale B., Rodier P.M.. Prenatal exposure of rats to valproic acid reproduces the cerebellar anomalies associated with autism. Neurotoxicol. Teratol. 2000; 22:319–324. [DOI] [PubMed] [Google Scholar]
- 16. Hsiao E.Y., McBride S.W., Hsien S., Sharon G., Hyde E.R., McCue T., Codelli J.A., Chow J., Reisman S.E., Petrosino J.F. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013; 155:1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith R.G., Kember R.L., Mill J., Fernandes C., Schalkwyk L.C., Buxbaum J.D., Reichenberg A.. Advancing paternal age is associated with deficits in social and exploratory behaviors in the offspring: a mouse model. PLoS One. 2009; 4:e8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Canetta S., Bolkan S., Padilla-Coreano N., Song L.J., Sahn R., Harrison N.L., Gordon J.A., Brown A., Kellendonk C.. Maternal immune activation leads to selective functional deficits in offspring parvalbumin interneurons. Mol. Psychiatry. 2016; 21:956–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller V.M., Zhu Y., Bucher C., McGinnis W., Ryan L.K., Siegel A., Zalcman S.. Gestational flu exposure induces changes in neurochemicals, affiliative hormones and brainstem inflammation, in addition to autism-like behaviors in mice. Brain Behav. Immun. 2013; 33:153–163. [DOI] [PubMed] [Google Scholar]
- 20. Margolis K.G., Li Z., Stevanovic K., Saurman V., Israelyan N., Anderson G.M., Snyder I., Veenstra-VanderWeele J., Blakely R.D., Gershon M.D.. Serotonin transporter variant drives preventable gastrointestinal abnormalities in development and function. J. Clin. Invest. 2016; 126:2221–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]