Abstract
Cancer is a genetic disease caused by somatic mutations; however, the understanding of the causative biological processes generating these mutations is limited. A cancer genome bears the cumulative effects of mutational processes during tumor development. Deciphering mutational signatures in cancer is a new topic in cancer research. The Wellcome Trust Sanger Institute (WTSI) has categorized 30 reference signatures in the COSMIC database based on the analyses of ∼10 000 sequencing datasets from TCGA and ICGC. Large cohorts and bioinformatics skills are required to perform the same analysis as WTSI. The quantification of known signatures in custom cohorts is not possible under the current framework of the COSMIC database, which motivates us to construct a database for mutational signatures in cancers and make such analyses more accessible to general researchers. mSignatureDB (http://tardis.cgu.edu.tw/msignaturedb) integrates R packages and in-house scripts to determine the contributions of the published signatures in 15 780 individual tumors from 73 TCGA/ICGC cancer projects, making comparison of signature patterns within and between projects become possible. mSignatureDB also allows users to perform signature analysis on their own datasets, quantifying contributions of signatures at sample resolution, which is a unique feature of mSignatureDB not available in other related databases.
INTRODUCTION
Cancer is a genetic disease that is caused by somatic mutations; however, the understanding of the causative biological processes of these mutations is limited. The catalogue of somatic mutations from a cancer genome bears the signatures of the mutational processes that have occurred during tumor development, which are the cumulative effects of the DNA damage and repair processes. Accordingly, deciphering these signatures in human cancer is a new trend in the cancer research community. Researchers from the Wellcome Trust Sanger Institute (WTSI) have analyzed somatic mutation spectra from over 7,000 cancers and revealed more than 20 distinct signatures using the algorithm of WTSI Mutational Signature Framework (1). This algorithm provides a better understanding of cancer biology by linking signatures to endogenous processes such as the enzymatic activity of DNA cytidine deaminases (APOBECs), the deficiency of DNA mismatch repair, or mutations in POLE and to exogenous mutagens such as tobacco, ultraviolet light and toxic chemicals.
Until now, 30 reference signatures were identified using the WTSI Mutational Signature Framework and have been categorized in the COSMIC database (2). Due to the ubiquitous nature of many of the signatures found across different cancer types, researchers may be interested in interrogating the presence and prevalence of published signatures in their collected tumor samples. As suggested in a previous study (3), at least 200 cancer genomes are required to decompose 20 signatures from their corresponding mutation catalogs, which means that large cohorts and adequate computing resources are necessary to perform the same analysis as WTSI. Additionally, the functionality of quantifying known signatures in small cohorts or single samples is not possible under the current WTSI framework.
To address the above issues, several R packages (4–6) were implemented to provide the computing infrastructure of the non-negative matrix factorization (NMF) algorithm, which is the core methodology of the WTSI Mutational Signature Framework used to decompose a mutation spectrum into signatures of biological processes. A recent development by Rachel et al. (7) has made possible the identification of mutational signatures within a single tumor sample, thus eliminating the need for large cohorts; however, most of the existing applications commonly lack the functionality to compare decomposed signatures to published signatures, which largely constrains their applicability. In addition, the existing database (2) only provides the signature distribution map across 40 cancer types (http://cancer.sanger.ac.uk/cosmic/signatures), detailed information such as the signature contributions in each cancer project or individual tumors, analysis framework for signature identification, and the search interface for comparing observed signatures with reference signatures are not available under current framework of the COSMIC database. Moreover, only half of the published signatures can be attributed to known mutational processes. Thus, an integrative database that comprehensively gathers mutational signature profiles of individual cancers as well as their corresponding clinical features may be beneficial for disclosing new connections between mutational processes and clinical features and identifying early diagnosis markers and potential therapeutic targets.
Our specific aim to construct mSignatureDB (http://tardis.cgu.edu.tw/msignaturedb) is to make mutational signature analyses more accessible to a wider community of researchers and to provide comprehensive insights into the common biological processes underlying the development of cancers. mSignatureDB integrates publicly available R packages and in-house scripts to determine the contributions of each published signature in 15 780 individual tumors from The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) across 73 cancer projects. User-friendly visualization options are provided to render the landscape of mutation signatures according to cancer types, projects, countries or clinical information, thereby facilitating in-depth analyses to better understand the etiology of mutational processes. Notably, mSignatureDB also accepts mutation profiles provided by users, ranging from a single tumor sample to a study cohort. The mutational signatures can be extracted from the uploaded mutation profile and assigned to reference signatures based on the NMF algorithm and cosine similarity method, respectively, making the comparison of the signature patterns within and between projects become possible, a unique feature of mSignatureDB that is currently not available in other related databases (2).
MATERIALS AND METHODS
Construction of mSignatureDB database
The current set of mutational signatures in mSignatureDB is based on an analysis of 15 780 tumors across 73 TCGA/ICGC cancer projects. Our purpose is to provide a database for users to compare mutational signatures with known mutation profiles collected from various resources, which is largely different from cBioPortal (8) that provides a web resource for exploring, visualizing, analyzing and downloading multidimensional cancer genomics data except for mutational signature analysis. The mutation profiles were downloaded from The National Cancer Institute (NCI) Genomic Data Commons (GDC) and the ICGC data portal data release 23 (https://dcc.icgc.org/releases/release_23/), along with their clinical information, including sex, lifestyle, patient history and tumor stage. The 30 published signatures categorized by different combination patterns of 96 tri-nucleotide mutation contexts were downloaded from COSMIC, which can function as a reference template for evaluating the degree of similarity between the observed and reference signatures (http://cancer.sanger.ac.uk/cancergenome/assets/signatures_probabilities.txt). For known signatures quantification, the R deconstructSigs package (7) was adopted to determine the composition of mutational signatures in individual tumor specimens of each TCGA/ICGC project as well as custom projects. For de novo and novel mutational signature analysis, the R mutSignatures package, an R-based implementation of the original WTSI Mutational Signature Framework (https://cran.r-project.org/web/packages/mutSignatures/index.html), was used to perform the NMF decomposition and estimate the stable number of decomposed signatures within each cancer project. To compare the observed signature with published signatures, the bootstrapped cosine similarity function implemented in the R supraHex package (9) was used to calculate statistical significance of the similarity between mutational signatures. The Maftools package (https://bioconductor.org/packages/release/bioc/html/maftools.html) was adopted to classify the somatic mutations into different substitution types to identify dominant mutation types as well as mutation hotspots along a reference genome. The g:Profiler package (10) was used to perform statistical enrichment analysis to identify over-represented gene ontology terms from top-ranked mutated genes associated with specific mutation types.
Architecture of mSignatureDB
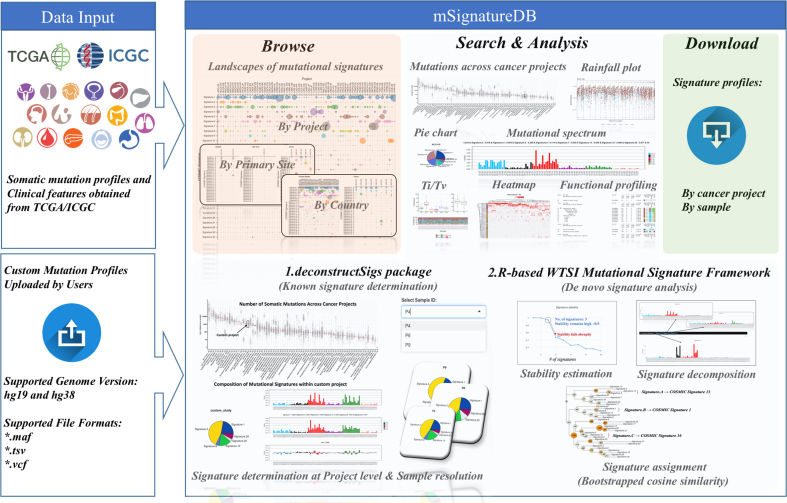
mSignatureDB includes three major components: (i) a web interface for inspection and retrieval of mutational signatures from a specific cancer project or individual tumors; (ii) a relational database implemented by R for storing the signature profiles from TCGA/ICGC tumors and their respective clinical information and (iii) a flexible framework for custom mutational signature analysis. The R shiny, plotly, rbokeh and canvasXpress packages were used to construct the interactive visualization framework of mSignatureDB (Figure 1). The custom signature analysis framework was linked to the Sun Grid Engine queuing system (11) to leverage the computationally intensive tasks.
Figure 1.
Overview of mSignatureDB. Somatic mutation profiles were gathered from TCGA/ICGC large-scale genomics studies. mSignatureDB comprises four components: (i) browse; (ii) search; (iii) analysis and (iv) download. In the ‘Browse’ page, the landscapes of mutational signatures can be inspected by cancer project, primary site or country. Users can search the database using the names of cancer projects. Hierarchically-clustered heamap is used to reveal dominant signatures in a cancer project according to the contribution of each signature. By displaying mutations according to substitution types and along a reference genome, users can easily depict dominant mutation types and localized regions of mutation hotspots. The signature profiles and the clinical associations can be downloaded through the ‘Download’ page. The web interfaces for two popular mutational signature analysis tools, the deconstructSigs and the WTSI Mutational Signature Framework, are provided to facilitate custom data analyses.
RESULTS AND DISCUSSION
Web interface
To facilitate the use of the mSignatureDB, we have established a user-friendly web interface to browse, search, and analyze mutational signatures from TCGA/ICGC projects as well as the custom data uploaded by users. The web interface comprises four main pages (Figure 1): (i) browse, (ii) search, (iii) analysis and (iv) download.
In the ‘Browse’ page, mSignatureDB integrated publicly available R packages and in-house scripts to determine the contributions of each published signature in 15 780 individual tumors across 73 ICGC cancer projects to provide an intuitive and efficient way for inspecting the signature landscape. Mutation signatures can be inspected through dot matrix and rendered according to their respective projects, primary sites and countries (http://tardis.cgu.edu.tw/msignaturedb/msignaturedb_help/browse.html#in-depth-understanding-of-the-landscapes-of-mutational-signatures-in-human-cancers). Detailed information about the signature contributions and their proposed etiologies can also be displayed through pop-up widows alongside the figures, eliminating the need for cross-referencing multiple websites. mSignatureDB can provide the most comprehensive roadmap describing the signatures of mutational processes operative in individual TCGA/ICGC tumors, which may be beneficial to explore different combinations of mutational signatures that are representative and distinct in specific cancer types or populations. We also provide a hyperlink to the COSMIC database for the convenience of inspecting the mutation pattern of each reference signature that is displayed as contribution of 96 trinucleotide contexts, to obtain a better understanding of a particular COSMIC signature.
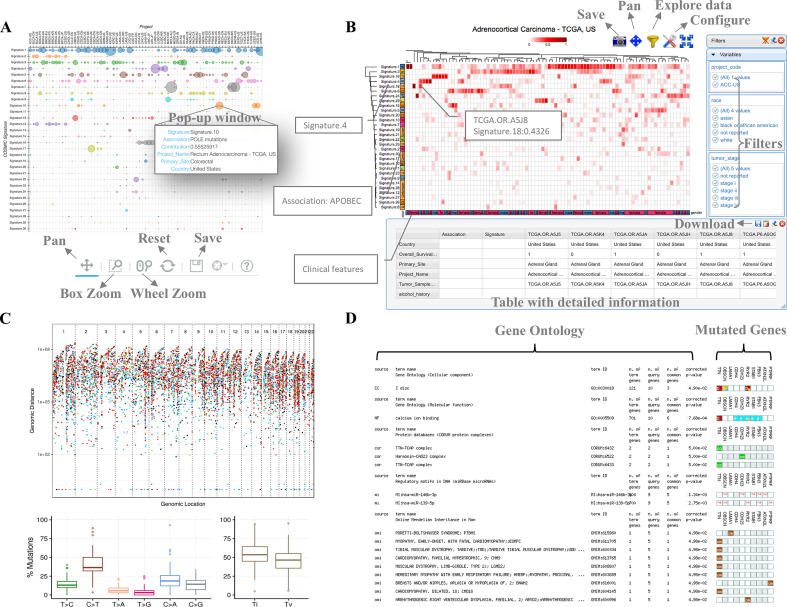
In the ‘Search’ page, the somatic mutation frequency, classification of SNV substitutions, mutation spectrum, and landscape of mutational signatures of each TCGA/ICGA project are searchable through a drop-down list with 73 cancer projects. The selected project will be highlighted in a summary plot about the number of somatic mutations across cancer projects worldwide along with its mutation spectrum and composition of mutational signatures. User-friendly control elements such as screen shot, zoom in/out, box select, auto scale and reset are available for the creation and manipulation of landscape maps of mutational signatures (Figure 2A). Users are able to select subsets of patients and compute statistically significant differences in signatures between clinical categories (e.g. sex, gender, vital status and tumor stage) on the fly with the aid of canvasXpress package. As shown in Figure 2B, the heatmap is used to perform a hierarchical clustering of samples based on the contribution of each mutational signature, which is helpful for depicting biomarkers for diagnostic, prognostic and therapeutic purposes (12). The rainfall plot is provided for facilitating detection of localized regions of hyper-mutation and identification of dominant mutation types (Figure 2C). Functional profiling of the most frequently mutated genes can be performed according to each mutation type to facilitate the users to identify their target of interests or potential therapeutic targets (Figure 2D).
Figure 2.
Output features of mSignatureDB. (A) Dot matrix is used to render the landscape of mutational signatures in each project. Explaining texts such as associated etiology and contribution of individual signatures are integrated in the plot and shown as pop-up windows. Flexible control elements are also available for the manipulation of the dot matrix. (B) Since the TCGA/ICGC mutation profiles and clinical information have been complied into mSignatureDB, users are able to compare mutational signatures between subsets of patients through the filters and the iterative heatmap. (C) Mutation hotspots are displayed as rainfall plot and box plots along a reference genome and according to substitution types, respectively. (D) Functional profiling of the most frequently mutated genes can be performed according to each substitution type to facilitate the users to identify their target of interests or potential therapeutic targets.
Due to the ubiquitous nature of the many signatures found across cancer types, researchers may be interested in knowing the presence and prevalence of these mutational signatures in their tumor samples. To make mutational signature analyses more accessible to general researchers, mSignatureDB provides user-friendly web interfaces for two popular mutational analysis tools, the deconstructSigs and the WTSI Mutational Signature Framework, and accepts mutation profiles in three different formats such as VCF, TSV (ICGC) and MAF (TCGA) from 2 different genome versions, which can be accessible through the ‘Analysis’ page of mSignatureDB (http://tardis.cgu.edu.tw/msignaturedb/Analysis/). Detailed descriptions about the input formats can be found on the tutorial page (http://tardis.cgu.edu.tw/msignaturedb/msignaturedb_help/analysis.html#mutational-signature-analysis-for-user-uploaded-data). The deconstructSigs approach is suitable for analyzing the contribution of known signatures in study cohorts with small number of samples, which can directly use published signatures as a reference, thus eliminating the need for large cohorts (3,7), and has been proved to be able to consistently identify the same signatures of mutational processes operative in a single tumor sample compared with the analysis of an entire sample set using the WTSI Mutational Signature Framework. However, most of the published signatures were identified by the WTSI Mutational Signature Framework, which is recommended for identifying novel mutational signatures when large samples are available. As suggested by previous study (3), at least 200 cancer genome catalogs are required for accurately decomposing signatures of 20 mutational processes. The original WTSI Mutational Signature Framework is developed on MATLAB, which requires a commercial license and basic knowledge on MALAB to perform de novo signature analysis. Accordingly, we incorporated the R-based implementation of the WTSI Mutational Signature Framework and provided a web interface for mSignatureDB users to simplify the analytical procedures. Signature assignment is the last step of de novo signature analysis, which can be achieved by cosine similarity analysis but always neglected by existing analysis packages. To facilitate known signature assignment, novel signature identification and give more confidence in the similarity analysis, the bootstrapped cosine similarity method is used to calculate statistical significance of similarity between mutational signatures. Furthermore, a bootstrapped tree (Figure 3) that summarizes the significance of similarity between mutational signatures is also provided to alleviate the exhausting and error-prone activity of visual inspection.
Figure 3.
Verification of Mutational Signature. The reference mutational signatures categorized in the COSMIC database were identified by the WTSI Mutational Signature Framework. Although the WTSI framework can perform de novo signature analysis and decompose signatures from mutation profiles, the signature assignment that can be achieved by cosine similarity analysis is always neglected by exiting tools, making the assignment of the decomposed signatures to published signatures very inconvenient. To address this issue and give more confidence in the similarity analysis, the bootstrapped cosine similarity method is used to calculate statistical significance of similarity between mutational signatures. As shown in this figure, a bootstrapped tree that summarizes the significance of cosine similarity between mutational signatures is provided to facilitate known signature assignment and novel signature identification while alleviate the exhausting and error-prone activity of visual inspection.
To obtain a better understanding of a particular COSMIC signature, detailed information about the composition of the 96 trinucleotide contexts and proposed etiology associated with each signature is provided as text files, which can be downloaded through the ‘Download’ page (http://tardis.cgu.edu.tw/msignaturedb/Download/).
Example of use
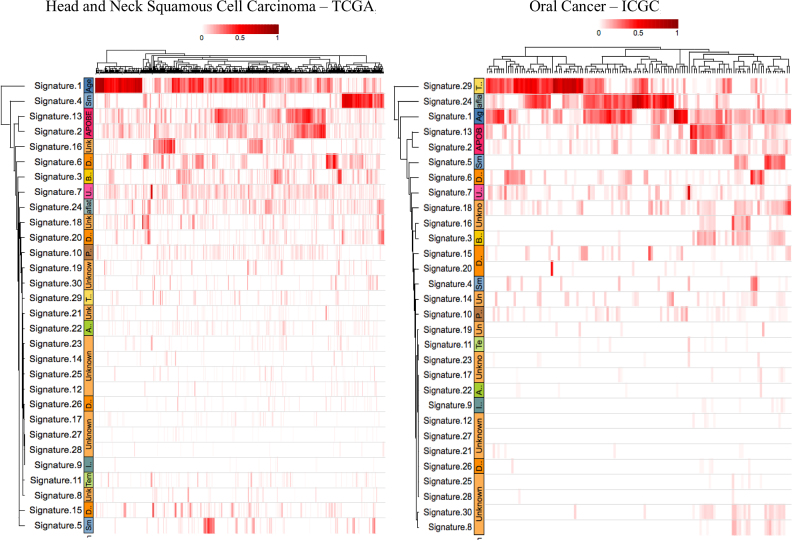
To illustrate and show an example of mSignatureDB functionalities, we have applied our application to analyze two public datasets reporting somatic mutation catalogs on 106 cases of oral squamous cell carcinomas (OSCC) from India (https://dcc.icgc.org/api/v1/download?fn=/release_23/Projects/ORCA-IN/simple_somatic_mutation.open.ORCA-IN.tsv.gz) and 510 cases of head and neck squamous cell cancer (HNSC) from America (https://dcc.icgc.org/api/v1/download?fn=/release_23/Projects/HNSC-US/sample.HNSC-US.tsv.gz), respectively. Because mSignatureDB can determine the composition of COSMIC reference signatures in individual tumor specimens, the signature landscapes can be easily compared at sample resolution within or across projects. Because the etiology of cancer is linked to several risk factors such as age, smoking, tobacco chewing, alcohol consumption, ultraviolet radiation and mutagen exposure, the samples can be further clustered into different subsets according to the contributions of mutational signatures and displayed as a hierarchically clustered heatmap. As shown in Figure 4, common signatures arise from aging (COSMIC signature 1), and the over-activity of APOBEC cytidine deaminases (COSMIC signature 2 and 13) can be identified through the heatmap. Furthermore, users can observe that smoking-related signature (COSMIC signature 4) plays key roles in the American population, whereas the tobacco chewing-related signature (COSMIC signature 29) is dominant in the Indian population. The result also demonstrates that risk factors can vary between countries and populations in head and neck cancer owing to different life habits or mutagen exposures. In addition to COSMIC signature 29, we noticed the prevalence of signature 24 in the Indian population, which is known to be correlated with aflatoxin exposure but was not emphasized by a previous publication (13). Detailed instructions on how to manipulate mSignatureDB to produce comparison heatmaps can refer to this link http://tardis.cgu.edu.tw/msignaturedb/msignaturedb_help/example-of-use.html.
Figure 4.
Example of Use. We have applied our application to analyze two public datasets reporting somatic mutation catalogs on 106 cases of OSCC from India and 510 cases of HNSC from America. Because mSignatureDB can determine the composition of COSMIC reference signatures in individual tumor specimens, signature landscapes can be compared at the sample resolution. As shown in this figure, signatures of active mutational processes such as aging (COSMIC signature 1) and over-activity of APOBEC enzymes (COSMIC signature 2 and 13) can be easily identified through the clustered heatmaps. Signatures originated from external mutagen exposures and habits (e.g. smoking and tobacco chewing) between different populations can also be identified using the visual analytic method.
CONCLUSIONS
Here, we present a database for deciphering mutational signatures in human cancers, mSignatureDB, which provides a portal for exploring the full landscape of mutational signatures present in 73 TCGA/ICGC cancer projects. As the sequencing of individual tumors becomes increasingly widely acceptable in a clinical setting, mSignatureDB also provides the ability to determine the composition of each COSMIC signature in individual samples from user-uploaded mutation profiles, thus making signature analysis more accessible to general users. mSignatureDB comprehensively gathers the mutational signature profiles of individual cancers alongside their corresponding clinical features from TCGA/ICGC cancer projects and has the potential benefits to identify early diagnosis markers, depict potential therapeutic targets and reveal new connections between mutational processes and clinical features. To the best of our knowledge, mSignatureDB is the only web application available as the most comprehensive source for categorizing known mutational signatures in cancer projects worldwide at a resolution of an individual sample.
ACKNOWLEDGEMENTS
We would like to thank members from Chang Gung Bioinformatics Core Laboratory for their comments and suggestions for this manuscript.
FUNDING
Chang Gung Memorial Hospital, Linkou [CMRPD1G0321/2, CMRPD1F0521/2/3]; Ministry of Science and Technology, Taiwan [MOST 104-2321-B-182–007-MY3, 105-2218-E-182-004, 106-2221-E-182-068]. Funding for open access charge: Chang Gung Memorial Hospital, Linkou, Taiwan [BMRP056] and Ministry of Science and Technology, Taiwan [MOST 104-2321-B-182-007-MY3].
Conflict of interest statement. None declared.
REFERENCES
- 1. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L. et al. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S. et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015; 43:D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alexandrov L.B., Nik-Zainal S., Wedge D.C., Campbell P.J., Stratton M.R.. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013; 3:246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gehring J.S., Fischer B., Lawrence M., Huber W.. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015; 31:3673–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ardin M., Cahais V., Castells X., Bouaoun L., Byrnes G., Herceg Z., Zavadil J., Olivier M.. MutSpec: a Galaxy toolbox for streamlined analyses of somatic mutation spectra in human and mouse cancer genomes. BMC Bioinformatics. 2016; 17:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gaujoux R., Seoighe C.. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010; 11:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rosenthal R., McGranahan N., Herrero J., Taylor B.S., Swanton C.. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016; 17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fang H., Gough J.. supraHex: an R/Bioconductor package for tabular omics data analysis using a supra-hexagonal map. Biochem. Biophys. Res. Commun. 2014; 443:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H., Vilo J.. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016; 44:W83–W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gentzsch W. Proceedings First IEEE/ACM International Symposium on Cluster Computing and the Grid. 2001; 35–36. [Google Scholar]
- 12. Harris R.S. Cancer mutation signatures, DNA damage mechanisms, and potential clinical implications. Genome Med. 2013; 5:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. India Project Team of the International Cancer Genome, C. Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat. Commun. 2013; 4:2873. [DOI] [PMC free article] [PubMed] [Google Scholar]