Abstract
Pharmaceuticals are designed to interact with specific molecular targets in humans and these targets generally have orthologs in other species. This provides opportunities for the drug discovery community to use alternative model species for drug development. It also means, however, there is potential for mode of action related effects in non-target wildlife species as many pharmaceuticals reach the environment through patient use and manufacturing wastes. Acquiring insight in drug target ortholog predictions across species and taxonomic groups has proven difficult because of the lack of an optimal strategy and because necessary information is spread across multiple and diverse sources and platforms. We introduce a new research platform tool, ECOdrug, that reliably connects drugs to their protein targets across divergent species. It harmonizes ortholog predictions from multiple sources via a simple user interface underpinning critical applications for a wide range of studies in pharmacology, ecotoxicology and comparative evolutionary biology. ECOdrug can be used to identify species with drug targets and identify drugs that interact with those targets. As such, it can be applied to support intelligent targeted drug safety testing by ensuring appropriate and relevant species are selected in ecological risk assessments. ECOdrug is freely accessible and available at: http://www.ecodrug.org.
INTRODUCTION
Drug discovery scientists screen and design pharmaceuticals based on their ability to interact with specific molecular targets in humans. The therapeutic dosing regimen is optimized for treatment efficacy while minimizing the risk of side effects caused by off-target interactions. The pharmaceutical industry invests huge resources into testing drugs, and non-human model species are an essential part of drug development, as many have the same targets. On the other hand, high evolutionary conservancy of drug targets means that upon release of pharmaceuticals to the environment, through patient use, manufacturing and inappropriate disposal of unused medicines, there is potential for therapeutic mode of action-related effects on wildlife species (1–3).
Understanding the conservation of human drug targets across species has applications in drug discovery, environmental protection and more generally in comparative physiology and evolutionary biology research. For example, in drug discovery the identification of appropriate orthologs in model species may allow for faster and more cost effective screening of drugs in a developmental pipeline (4–6). For any new market registration of medical products, in Europe and elsewhere (7,8), it is mandatory to perform an environmental risk assessment (ERA) which can involve environmental toxicological studies. Knowledge of drug target conservation can ensure that the most appropriate species are selected. In addition, by identifying taxonomic groups that lack a drug target unnecessary animal testing can be avoided.
To date, it has been difficult to acquire accurate insights on drug targets and their evolutionary conservation because the necessary information is not in a single repository and thus has to be retrieved from diverse sources. There are various datasets/databases that contain links between drugs and human protein targets e.g. Drugbank (9–11). But these sources do not cover the conservation of drug targets in a wide range of non-human species. Santos et al. (10) presents Ensembl ortholog predictions but this is limited to only 10 species. SeqAPASS is another resource that offers predictions of potential for chemicals to bind to targets in non-human species (12), but this system relies on less well developed methods for ortholog prediction and is password protected, limiting public access.
Establishing an evolutionary relationship between human drug target and non-human target is key as evolutionary related genes like orthologs are typically the most similar genes in the respective species in terms of sequence, structure and function (13)—providing the highest likelihood for a similar pharmacological effect to occur. Identification of orthologous relationships between genes is not a trivial undertaking as the true evolutionary relationships of genes are not known and need to be inferred from sequence data. There is no single strategy to predict orthology between human genes and those present in non-mammalian species and ortholog prediction is currently neither standardized nor optimized. Instead, a wide range of methods have been suggested, each with different strengths and weaknesses (14,15). Furthermore, predictions need to be updated on a regular basis, ideally concurrently with the release of new genomes and improvements in annotation of existing genomes as a static dataset will lose value over time.
Here we present ECOdrug (http://www.ecodrug.org), a user-friendly resource that provides an overview of innovative and legacy drugs, their human drug targets and the evolutionary conservation of these targets across taxonomic groups. It has been shown that combining of multiple ortholog prediction approaches can improve predictions of benchmarking datasets (16–18). Thus, to ensure an accurate prediction of orthologous genes, even across species that are evolutionary distant to one another, ECOdrug combines three well established methods for the ortholog predictions: Ensembl (19), EggNOG (20) and InParanoid (21). Importantly, we transparently show where the methods agree and disagree. The database is an integrated platform that allows for searches on drugs and/or drug targets and provides both high-level overviews of drug-target conservation and information of orthologs predicted in specific species. We provide examples of how ECOdrug can be used to select appropriate species for drug design or environmental tests and to facilitate the generation of testable hypotheses regarding pharmacologically related effects in wildlife species. We envisage that the database will provide a valuable resource for environmental toxicology, drug discovery and development, and safety communities by helping evaluate possibilities for read across of effect data from humans to wildlife and from model organisms to humans, as well as for research more widely for studies in comparative physiology and evolutionary biology.
DATABASE CONTENT
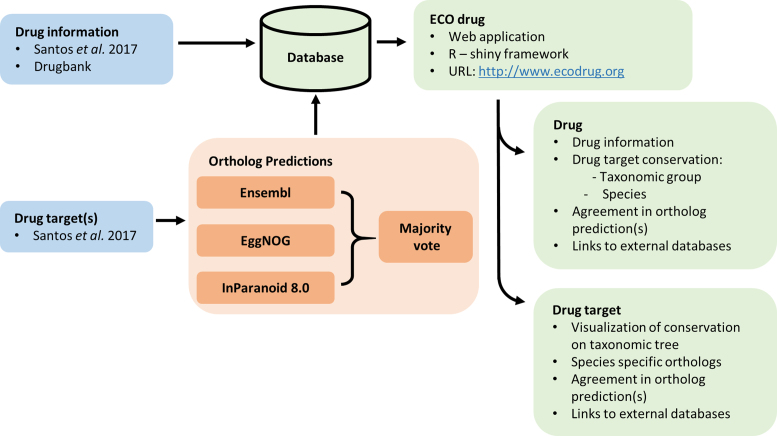
ECOdrug was built to provide a resource for obtaining information on drugs, their targets and associated ortholog predictions, currently for 640 eukaryotic species. The database combines data from multiple sources and illustrates the confidence of the ortholog predictions through a user friendly interface. An overview of the data sources and features of ECOdrug is shown in Figure 1.
Figure 1.
Schematic of the ECOdrug data sources and features. Information on drugs and associated targets (9) was combined with ortholog predictions (19–21) that can be accessed through the ECOdrug application.
Data collection
Pharmaceuticals and their associated drug targets
ECOdrug contains 1 194 Active Pharmaceutical Ingredients (APIs)—targeting 663 proteins sourced from the most recently published comprehensive map of molecular targets to which the approved drugs bind and are responsible for the therapeutic efficacy of the drug (10). The human target proteins are defined by their UniProt identifiers and we use biomaRt (22) to retrieve the associated Ensembl gene (n = 708) and Ensembl protein identifiers (n = 1 718). In addition to drug targets, the dataset also includes the following information on the APIs; therapeutic area, indicated by anatomical therapeutic chemical (ATC) codes (23), drug type (e.g. small molecule or protein) and associated mode(s) of action (10). Links to other databases such as DrugBank (9), UniProt (24) and Ensembl (19) enables users to quickly navigate to these sources, if desired.
Ortholog predictions
To obtain accurate ortholog predictions, ECOdrug aggregates predictions from three sources: Ensembl (19), EggNOG (20) and InParanoid (21). ECOdrug retrieves Ensembl orthologs by mapping ensembl gene IDs to all available ensembl homology attributes (data for first ECOdrug version, from February 2017). Beyond chordates the ortholog predictions are retrieved from Ensembl panCompara (ECOdrug initially uses release 30) (25). EggNOG ortholog predictions are retrieved by iterating over EggNOGs taxonomic levels from the closest to Homo sapiens (Hominidae NOG) to those most distant (Eukaryotes NOG). Orthologs to human drug targets in a given species were retrieved from the nearest possible taxonomic level. For InParanoid 8.0 (21) predictions we applied the standalone software package to derive orthologs between the Homo sapiens UniProt reference proteome (Initial ECOdrug predictions based on UP000005640_9606.fasta; UniProt Release 2016_04, Ensembl release 84 and Ensembl Genome release 31) and other reference proteomes available in the UniProt database. Sequence identity between drug targets and predicted orthologs were calculated using global alignment implemented in EMBOSS Needle (26). The ortholog predictions for Ensembl and EggNOG will be updated four times per year, while predictions for InParanoid are to be updated on an annual basis ensuring that the predictions will continue to be strengthened with new species, new gene builds and improved predictions.
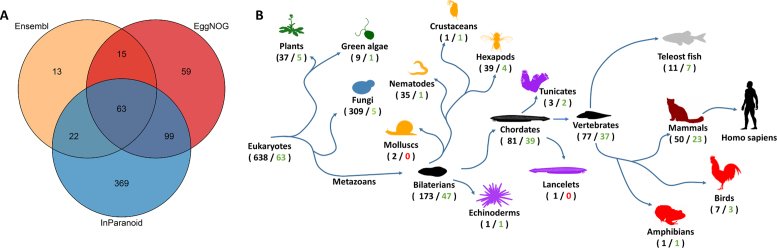
ECOdrug currently contains ortholog predictions for 640 species. However, species coverage differs for the available ortholog prediction methods. Predictions for 63 species derived from all three methods, and a further 136 species are represented in at least two prediction methods (Figure 2A). Mammals represent one third of the species that share predictions between the three methods, whereas some other taxonomic clades, e.g. molluscs, do not have any shared species (Figure 2B), reflecting both the significant knowledge gaps and the focus of the scientific community. For species present in multiple databases we combined predictions on a majority vote principle, i.e. two or more databases have to agree on the presence/absence of a drug target ortholog in a species. In cases where a species is represented in only two methods, majority vote calls presence of an ortholog if at least one database predicts an ortholog. The presentation in ECOdrug makes clear how many ortholog prediction methods support any given majority vote.
Figure 2.
Species coverage of predictions methods. (A) Each ortholog prediction method covers a different set of species. The overlap occurring between the methods are indicated by the numbers in the Venn diagram. (B) The taxonomic tree provides an overview of the species representation in the dataset. The number of species in taxonomic groups is indicated, with the numbers in black representing the total number of species, numbers in green illustrating the number of species shared among all three prediction methods and red is used where there are no shared species in the group. The colors of the species shades indicate larger taxonomic groups. Maroon/red: terrestrial vertebrates, gray: teleost fish, purple: invertebrate deuterostomes, orange: protostomes, blue: fungi, dark green: viridiplantae.
For each method the ortholog predictions are stored in .csv files, that can be downloaded through the ECOdrug application.
Patterns in drug target ortholog predictions
Agreement on ortholog presence and absence
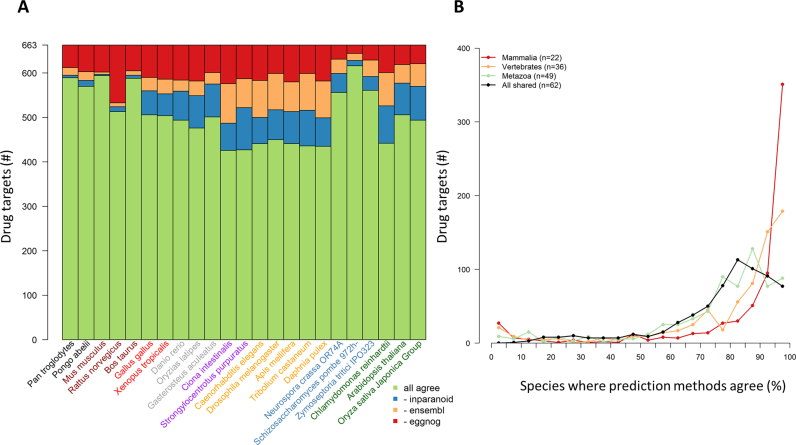
There was substantial agreement on the presence/absence of orthologs in species shared across the three databases (Figure 3A) with the tendency for increasing uncertainty for predictions with increasing taxonomic distance from humans. There was an agreement across the three databases for at least 76% of drug targets in vertebrates and around 65% for invertebrates, plants and algae. For fungi there was greater than an 83% agreement across the three databases which reflects the high quality fungal genome annotations and assemblies that have been developed by a large research community. The identity of drug targets with different results across the predictions methods varies across species and taxa. In other words, there is not a set of ‘contentious’ drug targets of which predictions of presence/absence of an ortholog in a species is always inconsistent across methods (Figure 3B).
Figure 3.
Agreement in ortholog predictions. (A) Bar colors indicate the agreement on presence/absence of drug target orthologs. The color of the species label indicates the taxonomic group (see Figure 2). (B) Histogram of the number of species with an ortholog prediction disagreement for drug targets. Circles indicate how many drug targets fit in each 5% bin. Line colors indicate which species are included in the histogram with only species shared by Ensembl, EggNOG and InParanoid being considered. Red: mammalian species, orange: vertebrate species, green: metazoan species, black: all shared species.
Drug target conservation patterns in taxonomic space
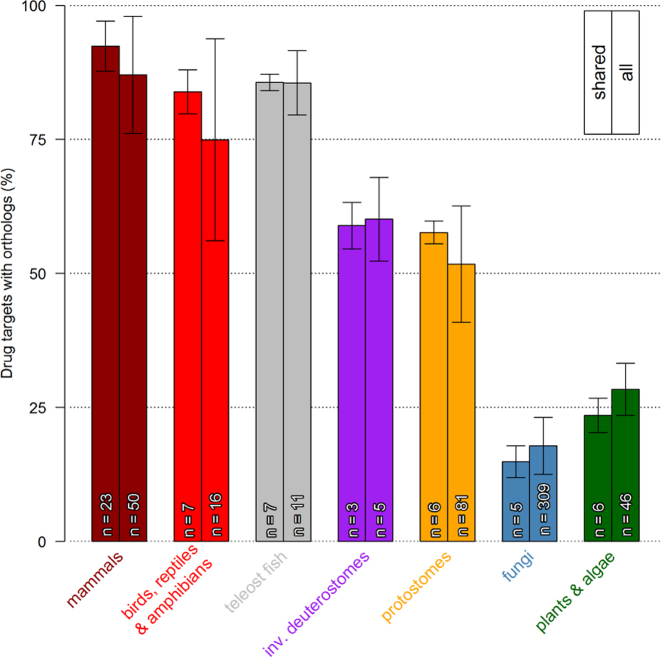
Mammalian species had the highest predicted number of human drug target orthologs, on average 92% of the targets had orthologs across the 23 species shared amongst all three prediction methods (Figure 4). Within the non-mammalian vertebrates (birds, reptiles and fish) there were similar levels of drug target conservation. In the invertebrate deuterostomes including the tunicate Ciona intestinalis and the sea urchin Strongylocentrotus purpuratus and in protostomes, there were orthologs to 50–65% of the human drug targets. Only the most evolutionary conserved genes might be expected to have orthologs in species in non-metazoan taxa such as fungi, plants and algae. In species in these taxa around 20–25% of the drug targets had predicted orthologs. This pattern of orthologs across the different taxa supports previously published results based on smaller scale assessments (10,27). We also analyzed drug target conservation across therapeutic space, showing that for drugs belonging to certain ATC classes conservation patterns are always identical (see Supplementary Materials File 1 and 2).
Figure 4.
Drug target conservation in taxonomic groups. Bars indicate the percentage of drug targets that have a predicted orthologs in a taxonomic group. Ortholog conservation and standard deviation are shown for both the shared species (left bar) and all species (right bar) in a taxonomic group.
FEATURES
The ECOdrug user interface includes one tab for drug related information and one tab for drug targets. The drug page (Supplementary Figure S2) has a search bar that allows searches for any drug name or DrugBank identifier. Users can view generic information on the drug, including a general description, drug type, ATC code and description of the mode of action. A table is provided that shows the drug targets of the selected drug and provides a high level overview of conservation for each drug target. In addition to the high level overview, a species level view of drug target conservation is available for all species, or a particular subset (e.g. only mammals). For majority vote presence/absence is indicated by TRUE/FALSE, whereas for individual databases the identifier (Ensembl gene id, Ensembl protein id or UniProt ID) and link to the external databases are provided. Sequence identities are displayed in brackets.
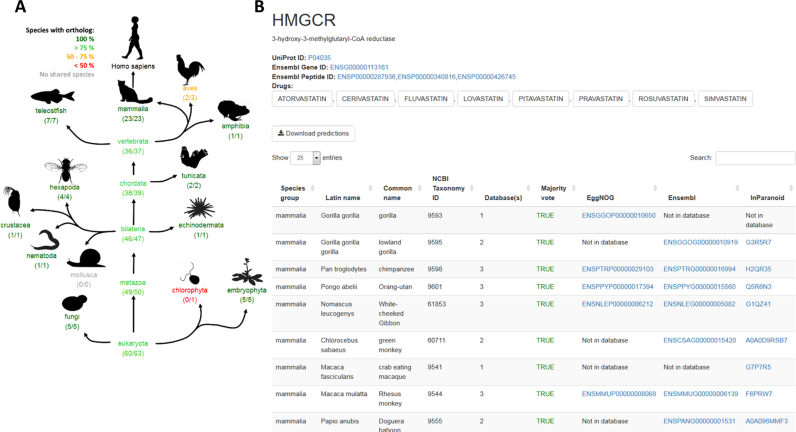
The drug target page (Figure 5) has a search functionality that accepts HGNC symbols, UniProt IDs, ensembl gene IDs and ensembl peptide IDs. The application will display a short description of the target, including links to all drugs that target the selected protein. An overview of the drug target conservation is visualized in a taxonomic tree, based on species shared by all three databases. In the same way as for the drug information page, the drug target page offers a table for species level predictions with Latin names, common names, NCBI taxonomy IDs, the number of databases wherein a species is represented and the majority vote/EggNOG/Ensembl/InParanoid predictions.
Figure 5.
ECODrug application: drug target page. The drug target page displays two sets of information for any given drug target. A graphic display of drug target conservation across taxonomic groups is shown in (A) with numbers based on species shared between all three prediction methods. Species specific ortholog predictions for combined and individual ortholog predictions methods are shown in a table (B).
CASE STUDIES
Here we provide three examples of the application of ECOdrug to: (i) identify appropriate model fish species for testing a drug class; (ii) selecting appropriate invertebrate test species for ERA and (iii) selecting appropriate species for the environmental protection of primary producers.
Case study I: identifying appropriate model fish species
Protein pump inhibitors such as Omeprazole, Dexlansoprazole and Esomeprazole are used in the therapy for stomach ulcers and act to block the H+/K+-ATPase proton pump (ATP4A) that is responsible the acidic conditions in the stomach. It has previously been shown that the presence of a gastric stomach phenotype correlates with the presence of ATP4A (28). In the first case study we applied ECOdrug to look for potential orthologs in fish for the target to proton pump inhibitors by selecting the drug target, ATP4A, and teleostei as the taxonomic group on the Drug Target page. The drug target page in ECOdrug illustrates that only 5 of the 11 fish teleost species listed have orthologs to ATP4A and these results agree with the pattern of conservation of ATP4A, as presented by Castro et al. (28) (see Supplementary Tables S1 and 2). Reliance on a single ortholog database, in this instance EggNOG, would have resulted in a false positive prediction as this database included a wider group of ATPase transporters as orthologs of ATP4A, reaffirming the merit of combining multiple prediction methods to best ensure accuracy.
Case study II: Selecting appropriate invertebrate test species for environmental risk assessment
The ERA of a human medical product within a marketing application requires tests on three species, representing different trophic levels. The invertebrate most commonly used is a daphnid species. Toxicity testing with Daphnia may, however, not be protective of other invertebrates if orthologs to the drug targets are absent in Daphnia but present in other invertebrate species. The ECOdrug database enables quick identification of such cases. To illustrate this, the ECOdrug predictions suggest that orthologs of the thyroid hormone receptors (THRA/THRB) are not present in Daphnia pulex, but do occur in some invertebrate deuterostomes (such as the sea urchin) and molluscs. These results align with a recent review on thyroid signaling conservation describing the presence of thyroid hormone receptors in invertebrate deuterostomes and nematodes—though they are not always responsive to thyroid hormone (29). In humans, defects in thyroid hormone signaling can lead to severe pathological conditions (30). The role of thyroid signaling in the invertebrates is not well understood and more research is required both to understand the basic biological process involved and thus the potential for drugs such as levothyroxine and liothyronine to affect invertebrates possessing orthologs of the thyroid hormone receptors.
Case study III: selecting appropriate species for environmental protection of primary producers
Cholesterol lowering drugs, also known as statins, are commonly prescribed and they include atorvastatin, lovastatin and rosuvastatin. These statins target 3-hydroxy-3-methylglutaryl Coenzyme A Reductase (HMG-CoA Reductase; HMGCR), the rate limiting enzyme in the mevalonate pathway, responsible for cholesterol synthesis. HMGCR is ubiquitous amongst eukaryotes and prokaryotes (31). In ERA primary producers are most often represented by a green algal species but results from ECOdrug indicate that in the case for statins other primary producers, e.g. plants, may be more appropriate for testing. ECOdrug illustrates that green algae species (chlorophytes) lack orthologs to HMGCR while higher plants have an HMGCR ortholog (Figure 5), a prediction that is in agreement with recent assessments of the molecular evolution of HMGCR (32). Further supporting this assumption, duckweed Lemna gibba has been reported to be at least 100 times more sensitive than green algae Chlamydomonas reinhardtii (24 000 μg/L) (33) to atorvastatin exposure (34). The toxicity for atorvastatin in L. gibba is in the same concentration range as its toxicity in daphnia (140 μg/L) and fish (450 μg/L) (33). Collectively, these results suggest that statins toxicity in plants is related to inhibition of HMGCR and indicate that toxicity tests in aquatic plants (or diatoms, or red algae) may be more protective of primary producers than tests with green algae.
FUTURE DIRECTIONS
We propose to further improve ECOdrug through integration with other platforms. First, with iPiEsys, a predictive framework that uses information from existing datasets on environmental fate and effects of APIs, toxicological studies, pharmacological mode of action and in silico models to support more intelligent environmental testing of pharmaceuticals. This is being developed by an Innovative Medicine Initiative EU consortium (http://i-pie.org/). Second, an initiative to develop evidence based tool to support the reduction, refinement and replacement of animals for experimentation (3Rs) for the ERA of pharmaceuticals is underway. The Virtual Fish Ecotoxicology Laboratory will use ECOdrug to help drive intelligent testing strategies that identify which fish species should be used for effect and bioaccumulation studies based on the presence of targets (http://www.simomics.com/). In addition, we aim to provide more detailed information regarding potential for binding of a drug to a drug target ortholog, for example in the form of binding site conservation. ECOdrug presents ortholog predictions for therapeutic drug targets only, but this can be expanded to include a wider set of proteins that are known to interact with drugs for example transporters and metabolising enzymes etc. The therapeutic score can also be widened to include antimicrobials and antiparasitics. Through integration with the systems outlined above, the addition of new features and regular updating we aim to ensure ECOdrug is maintained as a valuable and contemporary research tool for the communities in drug discovery, comparative and evolutionary biology and (eco)toxicology.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Adam Nellis and Paul Andrews from SimOmics for their help with deploying and hosting the application.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Innovative Medicines Initiative Joint Undertaking under Intelligent Assessment of Pharmaceuticals [115735]; European Union’s Seventh Framework Programme [FP7/2007–2013]; European Federation of Pharmaceutical Industries and Associations companies; FORMAS Research Council; Wallenberg Foundation; FRAM centre for future and chemical risk assessment and management strategies at University of Gothenburg; AstraZeneca (to J.R.S., S.F.O.) (in part); NC3Rs funded project [102519]; Virtual Fish EcoToxicology Laboratory; AstraZeneca Global SHE (to C.R.T., L.G., B.V.). Funding for open access charge: Innovative Medicines Initiative Joint Undertaking under Intelligent Assessment of Pharmaceuticals [115735]; European Union’s Seventh Framework Programme [FP7/2007–2013].
Conflict of interest statement. None declared.
REFERENCES
- 1. Boxall A.B.A., Rudd M.A., Brooks B.W., Caldwell D.J., Choi K., Hickmann S., Innes E., Ostapyk K., Staveley J.P., Verslycke T. et al. Pharmaceuticals and personal care products in the environment: what are the big questions?. Environ. Health Perspect. 2012; 120:1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Desbrow C., Routledge E.J., Brighty G.C., Sumpter J.P., Waldock M.. Identification of Estrogenic Chemicals in STW Effluent. 1. Chemical Fractionation and in Vitro Biological Screening. Environ. Sci. Technol. 1998; 32:1549–1558. [Google Scholar]
- 3. aus der Beek T., Weber F.-A., Bergmann A., Hickmann S., Ebert I., Hein A., Küster A.. Pharmaceuticals in the environment—global occurrences and perspectives. Environ. Toxicol. Chem. 2016; 35:823–835. [DOI] [PubMed] [Google Scholar]
- 4. Winter M.J., Redfern W.S., Hayfield A.J., Owen S.F., Valentin J.P., Hutchinson T.H.. Validation of a larval zebrafish locomotor assay for assessing the seizure liability of early-stage development drugs. J. Pharmacol. Toxicol. Methods. 2008; 57:176–187. [DOI] [PubMed] [Google Scholar]
- 5. Parker T., Libourel P.A., Hetheridge M.J., Cumming R.I., Sutcliffe T.P., Goonesinghe A.C., Ball J.S., Owen S.F., Chomis Y., Winter M.J.. A multi-endpoint in vivo larval zebrafish (Danio rerio) model for the assessment of integrated cardiovascular function. J. Pharmacol. Toxicol. Methods. 2014; 69:30–38. [DOI] [PubMed] [Google Scholar]
- 6. Zhang C., Ball J., Panzica-Kelly J., Augustine-Rauch K.. In vitro developmental toxicology screens: a report on the progress of the methodology and future applications. Chem. Res. Toxicol. 2016; 29:534–544. [DOI] [PubMed] [Google Scholar]
- 7. European Medicines Agency Guideline on the environmental risk assessment of medicinal products for human use. EMEA/CHMP/SWP/4447/00 corr. 2006; 2:4–12. [Google Scholar]
- 8. U.S. Department of Health and Human Services Food and Drug Administration Guidance for industry. Environmental assessment of human drug and biologics applications. CMC 6. 1998; 1–27. [Google Scholar]
- 9. Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014; 42:D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G., Karlsson A., Al-Lazikani B., Hersey A., Oprea T.I. et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017; 16:19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M., Krüger F.A., Light Y., Mak L., McGlinchey S. et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014; 42:D1083–D1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. LaLone C.A., Villeneuve D.L., Lyons D., Helgen H.W., Robinson S.L., Swintek J.A., Saari T.W., Ankley G.T.. Editor's highlight: sequence alignment to predict across species susceptibility (SeqAPASS): a web-based tool for addressing the challenges of cross-species extrapolation of chemical toxicity. Toxicol. Sci. 2016; 153:228–245. [DOI] [PubMed] [Google Scholar]
- 13. Gabaldon T., Koonin E.V.. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013; 14:360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kristensen D.M., Wolf Y.I., Mushegian A.R., Koonin E.V.. Computational methods for Gene Orthology inference. Brief. Bioinform. 2011; 12:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kriventseva E.V., Tegenfeldt F., Petty T.J., Waterhouse R.M., Simão F.A., Pozdnyakov I.A., Ioannidis P., Zdobnov E.M.. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015; 43:D250–D256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Altenhoff A.M., Boeckmann B., Capella-Gutierrez S., Dalquen D.A., DeLuca T., Forslund K., Huerta-Cepas J., Linard B., Pereira C., Pryszcz L.P.. Standardized benchmarking in the quest for orthologs. Nat. Methods. 2016; 13:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maher M.C., Hernandez R.D.. Rock, paper, scissors: harnessing complementarity in ortholog detection methods improves comparative genomic inference. G3. 2015; 5:629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pryszcz L.P., Huerta-Cepas J., Gabaldón T.. MetaPhOrs: orthology and paralogy predictions from multiple phylogenetic evidence using a consistency-based confidence score. Nucleic Acids Res. 2010; 39:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herrero J., Muffato M., Beal K., Fitzgerald S., Gordon L., Pignatelli M., Vilella A.J., Searle S.M., Amode R., Brent S.. Ensembl comparative genomics resources. Database. 2016; 2016:bav096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M.C., Rattei T., Mende D.R., Sunagawa S., Kuhn M.. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2015; 44:D296–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sonnhammer E.L., Östlund G.. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015; 43:D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Durinck S., Spellman P., Birney E., Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009; 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. WHO CollaboratingCentre for Drug Statistics Methodology Guidelines for ATC classification and DDD assignment 2013. 2013; Oslo: WHO; https://www.whocc.no/filearchive/publications/1_2013guidelines.pdf. [Google Scholar]
- 24. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kersey P.J., Lawson D., Birney E., Derwent P.S., Haimel M., Herrero J., Keenan S., Kerhornou A., Koscielny G., Kähäri A.. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010; 38:D563–D569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R.. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013; 41:W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gunnarsson L., Jauhiainen A., Kristiansson E., Nerman O., Larsson D.G.J.. Evolutionary conservation of human drug targets in organisms used for environmental risk assessments. Environ. Sci. Technol. 2008; 42:5807–5813. [DOI] [PubMed] [Google Scholar]
- 28. Castro L.F.C., Gonçalves O., Mazan S., Tay B.-H., Venkatesh B., Wilson J.M.. Recurrent gene loss correlates with the evolution of stomach phenotypes in gnathostome history. Proc. R. Soc. B Biol. Sci. 2014; 281:20132669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holzer G., Roux N., Laudet V.. Evolution of ligands, receptors and metabolizing enzymes of thyroid signaling. Mol. Cell. Endocrinol. 2017; doi:10.1016/j.mce.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 30. Pacini F., Castagna M.G., Brilli L., Pentheroudakis G.. Thyroid cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2012; 23(Suppl. 7):vii110–vii119. [DOI] [PubMed] [Google Scholar]
- 31. Friesen J.A., Rodwell V.W.. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biol. 2004; 5:248–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li W., Liu W., Wei H., He Q., Chen J., Zhang B., Zhu S.. Species-specific expansion and molecular evolution of the 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) gene family in plants. PLOS One. 2014; 9:e94172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vestel J., Caldwell D.J., Constantine L., D’Aco V.J., Davidson T., Dolan D.G., Millard S.P., Murray-Smith R., Parke N.J., Ryan J.J. et al. Use of acute and chronic ecotoxicity data in environmental risk assessment of pharmaceuticals. Environ Toxicol. Chem. 2016; 35:1201–1212. [DOI] [PubMed] [Google Scholar]
- 34. Brain R.A., Reitsma T.S., Lissemore L.I., Bestari K., Sibley P.K., Solomon K.R.. Herbicidal effects of statin pharmaceuticals in lemna gibba. Environ. Sci. Technol. 2006; 40:5116–5123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.