Abstract
Lnc2Meth (http://www.bio-bigdata.com/Lnc2Meth/), an interactive resource to identify regulatory relationships between human long non-coding RNAs (lncRNAs) and DNA methylation, is not only a manually curated collection and annotation of experimentally supported lncRNAs-DNA methylation associations but also a platform that effectively integrates tools for calculating and identifying the differentially methylated lncRNAs and protein-coding genes (PCGs) in diverse human diseases. The resource provides: (i) advanced search possibilities, e.g. retrieval of the database by searching the lncRNA symbol of interest, DNA methylation patterns, regulatory mechanisms and disease types; (ii) abundant computationally calculated DNA methylation array profiles for the lncRNAs and PCGs; (iii) the prognostic values for each hit transcript calculated from the patients clinical data; (iv) a genome browser to display the DNA methylation landscape of the lncRNA transcripts for a specific type of disease; (v) tools to re-annotate probes to lncRNA loci and identify the differential methylation patterns for lncRNAs and PCGs with user-supplied external datasets; (vi) an R package (LncDM) to complete the differentially methylated lncRNAs identification and visualization with local computers. Lnc2Meth provides a timely and valuable resource that can be applied to significantly expand our understanding of the regulatory relationships between lncRNAs and DNA methylation in various human diseases.
INTRODUCTION
Recent studies have identified massive of long RNA transcripts lacking of protein-coding potential, termed long non-coding RNAs (lncRNAs) (1). In view of the considerable diversity of lncRNAs and their involvement in important biological processes, numerous resources specific for lncRNAs have been developed. The primary resources mainly collect or integrate basic annotation and functional information on lncRNA transcripts, such as NONCODE (2), LNCipedia (3), and LNCat (4). Another type of resource lists functional lncRNAs that participate in disease, such as lncRNAdb (5), LncRNADisease (6) and Lnc2Cancer (7). The remaining resources explore the regulatory roles of lncRNAs interacting with other functional elements, such as genetic variants [LincSNP (8), lncRNASNP (9), and LncVar (10)], RNA editing sites [LNCediting (11)], microRNAs [DIANA-LncBase (12) and ChIPBase (13)] and protein-coding genes (PCGs) [LncRNA2Target (14) and LncReg (15)]. However, a resource linking DNA methylation, an essential epigenetic regulator and disease biomarker, with lncRNAs is still lacking.
DNA methylation is a fundamental feature of epigenomes that can affect the expression of protein-coding or non-coding transcripts (16,17). In addition to the direct regulation of lncRNAs by DNA methylation via interactions with their promoter regions, recent research has brought several more intricate regulatory relationships between lncRNAs and DNA methylation to light (18–21). For example, the lncRNA ecCEBPA (extra-coding CEBPA), transcribed from the CEBPA gene locus is reported to be critical for regulation of DNA methylation at this site through interactions with DNA methyltransferase 1, DNMT1 (22). Expression of the lncRNA maternally expressed gene 3 (MEG3), a well-characterized tumor inhibitor, is associated with its first intron methylation mediated by TET2 (23). However, the wealth of knowledge on lncRNA-methylation regulatory relationships is fragmented, with relevant research findings documented across thousands of different articles in the literature. Therefore, establishing a high-quality database with experimentally verified information on the regulatory relationships between lncRNAs and DNA methylation should greatly facilitate further research on the appropriate regulatory mechanisms and functions.
To address this gap, we developed a manually curated database, Lnc2Meth (http://www.bio-bigdata.com/Lnc2Meth/), with the aim of providing a comprehensive resource and web tool for clarifying the regulatory relationships between human lncRNAs and associated DNA methylation status. With the aid of Lnc2Meth, researchers can identify the lncRNAs dysmethylated in a specific disease or the diseases with a specific dysmethylated lncRNA. Furthermore, Lnc2Meth provides a platform that integrates tools to re-annotate probes from the Illumina Infinium Human Methylation 450k BeadChip (HM450k) array to lncRNA loci and identify differential methylation patterns of the lncRNAs and PCGs with user-supplied external datasets.
DATA COLLECTION AND DATABASE CONTENT
Collection of experimentally verified lncRNA-methylation associations
Manually curated lncRNA-methylation associations were systematically refined from the literature obtained by screening the PubMed database (before July 2017) with the following keywords combinations: (i) (long noncoding RNA or long non-coding RNA or long ncRNA or lncRNA or long intergenic noncoding RNA or long intergenic non-coding RNA or large intergenic noncoding RNA or large intergenic non-coding RNA or lincRNA) and (methylation or methylated or epigenetic or epigenetically); (ii) (lncRNA symbols or lncRNA alias or Ensembl gene IDs) and (methylation or methylated or epigenetic or epigenetically). We integrated 16 271 lncRNA symbols/aliases, and 15 693 Ensembl gene IDs, obtained from both lncRNAdb (5) and the Long non-coding RNA gene annotation file in GENCODE (Release 27) (24), as the keywords used in the literature-mining procedure. All selected studies were reviewed by at least two researchers. In this step, we retrieved the lncRNA symbol, transcript information (loci, type), DNA methylation region and pattern, experimental method, tissue/cell type, associated single nucleotide polymorphisms (SNPs)/microRNAs/mRNAs/proteins, disease type, prognostic value, literature reference (PubMed ID, year of publication, title of paper), a brief description of lncRNA and DNA methylation from the original studies, and the regulatory relationships.
Here, the regulatory relationships between lncRNAs and DNA methylation were categorized into three groups: Cis-Methylated LncRNAs (CML), Trans-Methylation Due to LncRNAs (TMDL) and Trans-Methylation Regulated LncRNAs (TMRL). In the CML group, DNA methylation adjacent to/on the lncRNAs loci, such as the promoter or the imprinting control region, directly modulates the expression of target lncRNAs as a cis-regulator (25,26). In the TMDL group, lncRNAs regulate the DNA methylation of a trans- genomic loci as an intermedium by recruiting the DNA (cytosine-5-)-methyltransferase (DNMT) (27,28). In the TMRL group, alteration of the DNA methylation state in specific genomic loci regulates the transcription of its antisense-oriented lncRNAs (29,30). Significantly, these types of regulatory processes are dysregulated in human cancers and implicated in disease progression (31). In addition, the method of disease classification in Lnc2Meth was based on the Disease Ontology database (DO; http://disease-ontology.org) (32). Comprehensive genome annotation files were obtained from GENCODE (https://www.gencodegenes.org/) (24). Furthermore, to facilitate access to information from external resources, we linked GeneCards (33), HGNC (34), Ensembl (35), NCBI GenBank (36), NONCODE (2), LNCipedia (37), lncRNAdb (5), LncRNAWiki (38), Lnc2Cancer (7), GREAT (39), OMIM (40) and COSMIC databases (41), which allowed the efficiently retrieval of a substantial amount of annotation and functional information from external resources.
Prediction of methylation patterns of lncRNAs
The HM450k array datasets were collected systematically from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/). The Whole-Genome Bisulfite Sequencing (WGBS) datasets were collected from the Encyclopedia of DNA Elements (ENCODE; https://www.encodeproject.org/) and TCGA. A re-annotation method was used to predict the methylation patterns for lncRNAs from these datasets (Supplementary Method). The information on the associations with lncRNAs expression status were calculated from the corresponding RNA-sequencing datasets collected from TCGA (Supplementary Method).
Database content and statistics
The current version of the Lnc2Meth documents 471 manually curated lncRNA-methylation associations by reviewing more than 3900 publicly published papers (Table 1). Lnc2Meth additionally provides 301 computationally calculated differential methylation profiles for lncRNAs and PCGs in 11 255 diseases and 1964 normal samples for 72 types of disease (Supplementary Figure S1A and B).
Table 1. Statistics of the LncRNA-Methylation associations.
| Regulatory category | Numbers of associations | Numbers of LncRNAs | Numbers of regulating partners/targets | Numbers of diseases | |||
|---|---|---|---|---|---|---|---|
| mRNAs | microRNAs | proteins | aSNPs | ||||
| aCML | 427 | 71 | 11 | 11 | 25 | 8 | 93 |
| aTMDL | 37 | 25 | 24 | 9 | 8 | — | 19 |
| aTMRL | 7 | 6 | 1 | — | — | — | 4 |
| Total | 471 | 95 | 36 | 20 | 30 | 8 | 99 |
aSNP is short for single nucleotide polymorphism. CML is short for cis-methylated LncRNA. TMDL is short for trans-methylation due to LncRNA. TMRL is short for trans-methylation regulated LncRNA.
Database implementation
The database was organized with MySQL (version 5.6.25) and queried using JSP scripts. The web interface was developed using HTML with JavaScript. The ‘DMBrowser’ module was constructed with JBrowse (release 1.12.1) to navigate transcript structure and explore the methylation patterns (42).
DATABASE FEATURES AND APPLICATIONS
Web interface
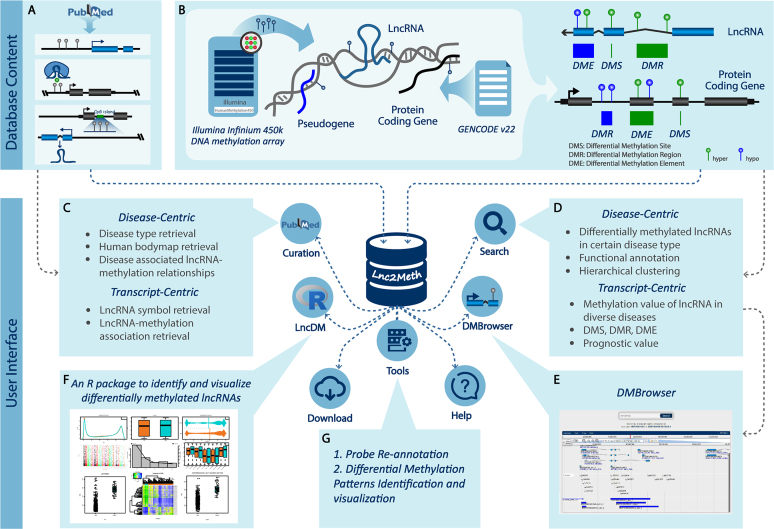
We provided a user-friendly web interface (Figure 1) for users to query the database through multiple modules and identify differential methylation patterns for a given lncRNA including (i) ‘Curation’, a retrieval module for experimentally verified lncRNA-methylation associations (Figure 2A–C, Supplementary Method), (ii) ‘Search’, a retrieval module for predicted differential methylation patterns of lncRNAs and PCGs (Figure 2D–K, Supplementary Method), (iii) ‘DMBrowser’, a genome browser for illustrating the methylation landscape of lncRNAs and PCGs, (iv) ‘Tools’, a server for identifying the differential methylation patterns of lncRNAs and PCGs online, (v) ‘LncDM’, an R package for calculating differential methylation patterns of lncRNAs and PCGs on local computers, (vi) ‘Download’, a module for downloading the differential methylation pattern profiles of lncRNAs and PCGs and (vii) ‘Help’, a module with detailed documentation of user tutorials.
Figure 1.
Content and interface of Lnc2Meth. (A) Manually curated lncRNA-methylation associations in Lnc2Meth. (B) Predicted differential methylation patterns of lncRNAs and PCGs in Lnc2Meth. (C) Curation module for manually curated lncRNA-methylation associations. (D) Search module for predicted lncRNA/PCG methylation patterns. (E) DMBrowser module for illustrating methylation landscape of lncRNAs and PCGs. (F) LncDM, an R package for calculating methylation patterns of lncRNAs and PCGs on local computers. (G) Tools module for identifying the differential methylation patterns of lncRNAs and PCGs online.
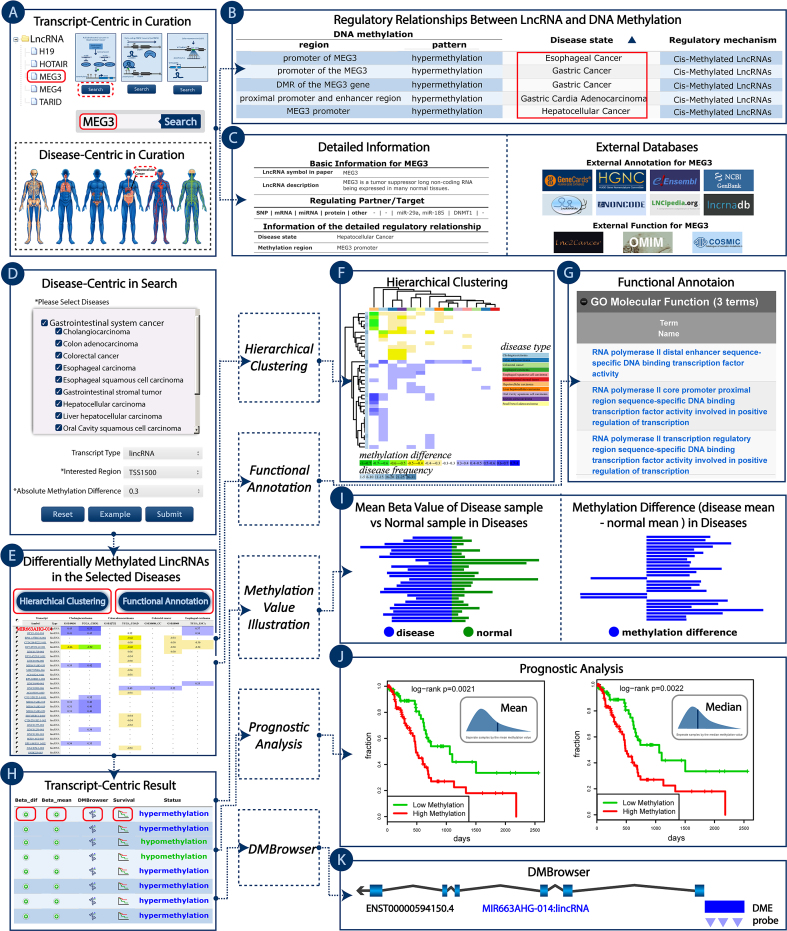
Figure 2.
Case study of using Lnc2Meth. (A) The interface of the curation module, with MEG3 input or selected as the retrieved lncRNA in ‘Transcript-Centric’ page. (B) Search result page of MEG3. (C) Search result page with detailed information. (D) The ‘Disease-Centric’ page of the search module with ‘gastrointestinal system cancer’ as ‘Select Disease’, ‘lincRNA’ as ‘Transcript Type’, ‘TSS1500’ as ‘Interested Region’, ‘0.3’ as ‘Absolute Methylation Difference’. (E) Search result page of ‘gastrointestinal system cancer’. (F) Hierarchical clustering heat map of the methylation profile for selected lincRNAs. (G) Functional annotation results of the selected lincRNAs using GREAT. (H) Search result page of MIR663AHG-014. (I) Bar plots to illustrate the differentially methylated MIR663AHG-014 in diseases. (J) Two Kaplan–Meier curves for MIR663AHG-014 in Pancreatic adenocarcinoma. (K) DNA methylation landscape of MIR663AHG-014 in Lnc2Meth DMBrowser.
Online tools for probe re-annotation and identification of differential methylation patterns for lncRNA/PCG’s
Lnc2Meth provides two online tools. One is the ‘Probe Re-annotation’ tool (PR) and the other is the ‘Differential Methylation Identification’ tool (DMI). With the PR, users could obtain the annotation information for the HM450k array probes located in the functional regions (TSS200, TSS1500, 1_exon, intron, gene body, TSS10kb and TTS10kb) of their interested lncRNA/PCG by searching the gene symbol, Ensembl gene ID or the genomic loci. With the DMI, users could get the calculating results of the differential methylation patterns for their inputting lncRNA/PCG by uploading the user-supplied external HM450k array datasets (Supplementary Figure S2, Supplementary Method). Except for the lncRNAs with known gene symbol or Ensembl Gene ID, users could also re-annotate and calculate methylation patterns for a newly-assembled lncRNA by providing its genomic Loci. These analysis tools allow users to mine their own data to determine whether a given lncRNA is dysmethylated between the case and control samples.
DMBrowser of Lnc2Meth
JBrowse (release 1.12.1) was applied to develop a genome browser, DMBrowser, to provide a user-friendly interface for navigating the transcript structure and visualizing the differential methylation patterns of the transcript in specific diseases. Users could browse the structures and loci of the lncRNAs and the re-annotated probes by submitting the lncRNAs symbol, transcript ID or genomic interval. Tracks of methylation patterns in diverse diseases could be selected and added into the ‘Search’ page. The hit transcripts from the transcript-centric search module also contain links to DMBrowser. For the track of lncRNA transcripts, DMBrowser provides links to the UCSC Genome Browser for users further investigating the characteristics of the lncRNAs and methylation.
DISCUSSION AND FUTURE EXTENSIONS
In this study, we developed a database aimed to collect and illustrate the regulatory relationships between lncRNAs and DNA methylation with both experimentally verified and predicted information. Existing methylation-related databases that are widely in use have mainly focused on PCG and seldom contain lncRNA records (Supplementary Table S1). For example, PubMeth (43), MeInfoText (44) and DDMGD (45) amass data on gene-centric methylation data in disease while DiseaseMeth (46) and MethyCancer (47) provided information on disease-associated alterations of DNA methylation patterns with scattered records of lncRNAs. Therefore, Lnc2Meth, a resource for identifying DNA methylation associations with the lncRNAs, represents an early step towards meeting the extensive research interest and should facilitate the generation and systematic analysis of novel hypotheses regarding the regulatory mechanisms of lncRNA-DNA methylation associations.
In the future, we will continue to update and integrate data content in Lnc2Meth by: (i) continuous literature mining and information refining, (ii) expanding the available DNA methylation datasets detected by a variety of techniques, such as the HM450k array, WGBS, reduced representation bisulfite sequencing, and MeDIP-Seq. In addition, we plan to revise information on lncRNA annotation as an improvement of the GENCODE gene annotation catalogues. Lnc2Meth will be maintained and updated to ensure that it remains a useful resource for the research community.
Supplementary Material
ACKNOWLEDGEMENTS
We thank TCGA, GEO, GENCODE, ENCODE and other projects for generating and sharing the data used in this paper. We acknowledge Lining Zhang and Jing Gan for their valuable suggestion on data curation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Natural Science Foundation of China [31501038, 91439117, 61473106, 31401090, 31601080]; Research Fund for the Postdoctoral Science Foundation of China [2016M591546]; Heilongjiang Province [LBH-Z15132]; National High Technology Research and Development Program of China [863 Program, 2014AA021102]; National Program on Key Basic Research Project [973 Program, 2014CB910504]; Yu Weihan Outstanding Youth Training Fund of Harbin Medical University. Funding for open access charge: National Natural Science Foundation of China [31501038, 91439117, 61473106, 31401090, 31601080].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kung J.T., Colognori D., Lee J.T.. Long noncoding RNAs: past, present, and future. Genetics. 2013; 193:651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q. et al. . NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016; 44:D203–D208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P.. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015; 43:D174–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu J., Bai J., Zhang X., Lv Y., Gong Y., Liu L., Zhao H., Yu F., Ping Y., Zhang G. et al. . A comprehensive overview of lncRNA annotation resources. Brief. Bioinform. 2017; 18:236–249. [DOI] [PubMed] [Google Scholar]
- 5. Quek X.C., Thomson D.W., Maag J.L., Bartonicek N., Signal B., Clark M.B., Gloss B.S., Dinger M.E.. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015; 43:D168–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q.. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013; 41:D983–D986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ning S., Zhang J., Wang P., Zhi H., Wang J., Liu Y., Gao Y., Guo M., Yue M., Wang L. et al. . Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016; 44:D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ning S., Yue M., Wang P., Liu Y., Zhi H., Zhang Y., Zhang J., Gao Y., Guo M., Zhou D. et al. . LincSNP 2.0: an updated database for linking disease-associated SNPs to human long non-coding RNAs and their TFBSs. Nucleic Acids Res. 2017; 45:D74–D78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gong J., Liu W., Zhang J., Miao X., Guo A.Y.. lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2015; 43:D181–D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen X., Hao Y., Cui Y., Fan Z., He S., Luo J., Chen R.. LncVar: a database of genetic variation associated with long non-coding genes. Bioinformatics. 2017; 33:112–118. [DOI] [PubMed] [Google Scholar]
- 11. Gong J., Liu C., Liu W., Xiang Y., Diao L., Guo A.Y., Han L.. LNCediting: a database for functional effects of RNA editing in lncRNAs. Nucleic Acids Res. 2017; 45:D79–D84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paraskevopoulou M.D., Vlachos I.S., Karagkouni D., Georgakilas G., Kanellos I., Vergoulis T., Zagganas K., Tsanakas P., Floros E., Dalamagas T. et al. . DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016; 44:D231–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou K.R., Liu S., Sun W.J., Zheng L.L., Zhou H., Yang J.H., Qu L.H.. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017; 45:D43–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang Q., Wang J., Wu X., Ma R., Zhang T., Jin S., Han Z., Tan R., Peng J., Liu G. et al. . LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res. 2015; 43:D193–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou Z., Shen Y., Khan M.R., Li A.. LncReg: a reference resource for lncRNA-associated regulatory networks. Database (Oxford). 2015; 2015:bav083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Borrello M.G., Pierotti M.A., Tamborini E., Biassoni D., Rizzetti M.G., Pilotti S., Della Porta G.. DNA methylation of coding and non-coding regions of the human H-RAS gene in normal and tumor tissues. Oncogene. 1992; 7:269–275. [PubMed] [Google Scholar]
- 17. Li D., Da L., Tang H., Li T., Zhao M.. CpG methylation plays a vital role in determining tissue- and cell-specific expression of the human cell-death-inducing DFF45-like effector A gene through the regulation of Sp1/Sp3 binding. Nucleic Acids Res. 2008; 36:330–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhi H., Ning S., Li X., Li Y., Wu W., Li X.. A novel reannotation strategy for dissecting DNA methylation patterns of human long intergenic non-coding RNAs in cancers. Nucleic Acids Res. 2014; 42:8258–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma X., Yu L., Wang P., Yang X.. Discovering DNA methylation patterns for long non-coding RNAs associated with cancer subtypes. Comput. Biol. Chem. 2017; 69:164–170. [DOI] [PubMed] [Google Scholar]
- 20. Bohmdorfer G., Rowley M.J., Kucinski J., Zhu Y., Amies I., Wierzbicki A.T.. RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J. 2014; 79:181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li W., Zheng J., Deng J., You Y., Wu H., Li N., Lu J., Zhou Y.. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014; 146:1714–1726. [DOI] [PubMed] [Google Scholar]
- 22. Di Ruscio A., Ebralidze A.K., Benoukraf T., Amabile G., Goff L.A., Terragni J., Figueroa M.E., De Figueiredo Pontes L.L., Alberich-Jorda M., Zhang P. et al. . DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013; 503:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lyu Y., Lou J., Yang Y., Feng J., Hao Y., Huang S., Yin L., Xu J., Huang D., Ma B. et al. . Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia. 2017; doi:10.1038/leu.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. . GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bohne F., Langer D., Martine U., Eider C.S., Cencic R., Begemann M., Elbracht M., Bulow L., Eggermann T., Zechner U. et al. . Kaiso mediates human ICR1 methylation maintenance and H19 transcriptional fine regulation. Clin. Epigenet. 2016; 8:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rokavec M., Horst D., Hermeking H.. Cellular model of colon cancer progression reveals signatures of mRNAs, miRNA, lncRNAs, and epigenetic modifications associated with metastasis. Cancer Res. 2017; 77:1854–1867. [DOI] [PubMed] [Google Scholar]
- 27. Vennin C., Spruyt N., Robin Y.M., Chassat T., Le Bourhis X., Adriaenssens E.. The long non-coding RNA 91H increases aggressive phenotype of breast cancer cells and up-regulates H19/IGF2 expression through epigenetic modifications. Cancer Lett. 2017; 385:198–206. [DOI] [PubMed] [Google Scholar]
- 28. Yu F., Chen B., Dong P., Zheng J.. HOTAIR Epigenetically Modulates PTEN expression via MicroRNA-29b: a novel mechanism in regulation of liver fibrosis. Mol. Ther. 2017; 25:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao T., He B., Pan Y., Xu Y., Li R., Deng Q., Sun H., Wang S.. Long non-coding RNA 91H contributes to the occurrence and progression of esophageal squamous cell carcinoma by inhibiting IGF2 expression. Mol. Carcinog. 2015; 54:359–367. [DOI] [PubMed] [Google Scholar]
- 30. McCarty G., Loeb D.M.. Hypoxia-sensitive epigenetic regulation of an antisense-oriented lncRNA controls WT1 expression in myeloid leukemia cells. PLoS One. 2015; 10:e0119837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lai F., Shiekhattar R.. Where long noncoding RNAs meet DNA methylation. Cell Res. 2014; 24:263–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kibbe W.A., Arze C., Felix V., Mitraka E., Bolton E., Fu G., Mungall C.J., Binder J.X., Malone J., Vasant D. et al. . Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015; 43:D1071–D1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H. et al. . GeneCards Version 3: the human gene integrator. Database (Oxford). 2010; 2010:baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yates B., Braschi B., Gray K.A., Seal R.L., Tweedie S., Bruford E.A.. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017; 45:D619–D625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T. et al. . The Ensembl genome database project. Nucleic Acids Res. 2002; 30:38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karsch-Mizrachi I., Ouellette B.F.. The GenBank sequence database. Methods Biochem. Anal. 2001; 43:45–63. [PubMed] [Google Scholar]
- 37. Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P.. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015; 43:4363–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma L., Li A., Zou D., Xu X., Xia L., Yu J., Bajic V.B., Zhang Z.. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015; 43:D187–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feramisco J.D., Sadreyev R.I., Murray M.L., Grishin N.V., Tsao H.. Phenotypic and genotypic analyses of genetic skin disease through the Online Mendelian Inheritance in Man (OMIM) database. J. Invest. Dermatol. 2009; 129:2628–2636. [DOI] [PubMed] [Google Scholar]
- 41. Bamford S., Dawson E., Forbes S., Clements J., Pettett R., Dogan A., Flanagan A., Teague J., Futreal P.A., Stratton M.R. et al. . The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer. 2004; 91:355–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L. et al. . JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ongenaert M., Van Neste L., De Meyer T., Menschaert G., Bekaert S., Van Criekinge W.. PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucleic Acids Res. 2008; 36:D842–D846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fang Y.C., Lai P.T., Dai H.J., Hsu W.L.. MeInfoText 2.0: gene methylation and cancer relation extraction from biomedical literature. BMC Bioinformatics. 2011; 12:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bin Raies A., Mansour H., Incitti R., Bajic V.B.. DDMGD: the database of text-mined associations between genes methylated in diseases from different species. Nucleic Acids Res. 2015; 43:D879–D886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiong Y., Wei Y., Gu Y., Zhang S., Lyu J., Zhang B., Chen C., Zhu J., Wang Y., Liu H. et al. . DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res. 2017; 45:D888–D895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. He X., Chang S., Zhang J., Zhao Q., Xiang H., Kusonmano K., Yang L., Sun Z.S., Yang H., Wang J.. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res. 2008; 36:D836–D841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.