Abstract
The GWIPS-viz browser (http://gwips.ucc.ie/) is an on-line genome browser which is tailored for exploring ribosome profiling (Ribo-seq) data. Since its publication in 2014, GWIPS-viz provides Ribo-seq data for an additional 14 genomes bringing the current total to 23. The integration of new Ribo-seq data has been automated thereby increasing the number of available tracks to 1792, a 10-fold increase in the last three years. The increase is particularly substantial for data derived from human sources. Following user requests, we added the functionality to download these tracks in bigWig format. We also incorporated new types of data (e.g. TCP-seq) as well as auxiliary tracks from other sources that help with the interpretation of Ribo-seq data. Improvements in the visualization of the data have been carried out particularly for bacterial genomes where the Ribo-seq data are now shown in a strand specific manner. For higher eukaryotic datasets, we provide characteristics of individual datasets using the RUST program which includes the triplet periodicity, sequencing biases and relative inferred A-site dwell times. This information can be used for assessing the quality of Ribo-seq datasets. To improve the power of the signal, we aggregate Ribo-seq data from several studies into Global aggregate tracks for each genome.
INTRODUCTION
Ribosome profiling (Ribo-seq) is a biochemical technique that utilizes high throughput sequencing that captures the mRNA fragments that are protected by actively translating ribosomes (1) thereby providing Genome-Wide Information on Protein Synthesis (GWIPS) (2). Ribo-seq was first carried out in Saccharomyces cerevisiae (1) and has since been used in many organisms resulting in a substantial growth in the number of published datasets. The numerous applications of the ribosome profiling technique as well as its limitations are described in details elsewhere (3–14). While the majority of Ribo-seq datasets represent footprints of elongating ribosomes, a number of studies have used protocols for enriching footprints deriving from initiating ribosomes and more recently a modification of the ribosome profiling protocol allowed footprinting of scanning ribosomes (15).
To account for differences in mRNA abundance, most Ribo-seq studies also generate parallel datasets where total mRNA (or total RNA) is randomly degraded and subsequently sequenced. Here we refer to such datasets as mRNA-seq. To date, the majority of published Ribo-seq/mRNA-seq raw sequencing data have been deposited in NCBI’s Sequence Read Archive (SRA) (16).
The GWIPS-viz browser (http://gwips.ucc.ie/) uses the functionality of the UCSC Genome Browser (17) to provide visualizations of Ribo-seq coupled with mRNA-seq controls so that users can freely explore pre-populated Ribo-seq/mRNA-seq tracks without the need to download, pre-process and align raw sequencing data to the corresponding genomes. Since its original publication (18), we have striven to expand the repertoire of Ribo-seq/mRNA-seq data hosted on GWIPS-viz. We have also incorporated additional tracks as well as improved visualizations to help users better interpret the Ribo-seq/mRNA-seq data.
New genomes in GWIPS-viz
In 2014, GWIPS-viz provided Ribo-seq/mRNA-seq data for nine genomes: Homo sapiens (hg19), Mus musculus (mm10), Danio rerio (danRer7), Caenorhabditis elegans (ce10), S. cerevisiae (sacCer3), Escherichia coli K12 (ASM584_v2), Bacillus subtilis (11/09/2009), human cytomegalovirus (HHV5 strain Merlin) and bacteriophage lambda (NC_001416). Today GWIPS-viz provides Ribo-seq/mRNA-seq data for an additional 14 genomes: Rattus novegicus (rn6), Xenopus laevis (v6.0), Drosophila melanogaster (dm3), Trypanosoma brucei brucei (TriTrypDb TREU927 – v 5.1), Plasmodium falciparum (ASM276v1), Schizosaccharomyces pombe (ASM294v2), Neurospora crassa (or74a/GCF_000182925.2_NC12), Arabidopsis thaliana (Nov-2013), Zea Mays B73 (GCF_000005005.1_NC_024459.1), E. coli BW25113 (ASM75055v1), Caulobacter crescentus (ASM2200v1), Streptomyces coelicolor (ASM20383v1), Staphylococcus aureus USA300_FPR3757 (ASM1346 v1), S. aureus NCTC 8325 (ASM1342 v1). In addition, the more recent hg38 version of the human genome assembly has been provided.
New tracks in GWIPS-viz
As well as the addition of new genomes to GWIPS-viz, the number of hosted tracks has grown by 10-fold. This is largely a result of our automated computational pipeline for the integration of new Ribo-seq and mRNA-seq data for genomes already in the browser, bringing the total number of tracks to 1792 tracks across the 23 genomes. The increase has been particularly substantial for Ribo-seq data generated for human as well as for mouse and S. cerevisiae. New data since the original GWIPS-viz publication include: H. sapiens hg38 assembly (19–49), H. sapiens hg19 assembly (43,50–54), M. musculus (51,52,55–75), R. novegicus (76–78), D. rerio (79–81), X. laevis (82), C. elegans (83–85), D. melanogaster (86–88), T. brucei brucei (89), P. falciparum (90), S. cerevisiae (15,64,82,85,91–109), S. pombe (82,110), N. crassa (111), A. thaliana (112–114), Z. Mays B73 (115), E. coli K12 (116–124), E. coli BW25113 (125), B. subtilis (126), C. crescentus (127), S. coelicolor (128), S. aureus USA300_FPR3757 (129), S. aureus NCTC 8325 (130). This expansion of datasets allows for improved cross-species comparison of orthologous genes while the availability of datasets from multiple research groups permits the assessment of technical reproducibility of the ribosome densities (131).
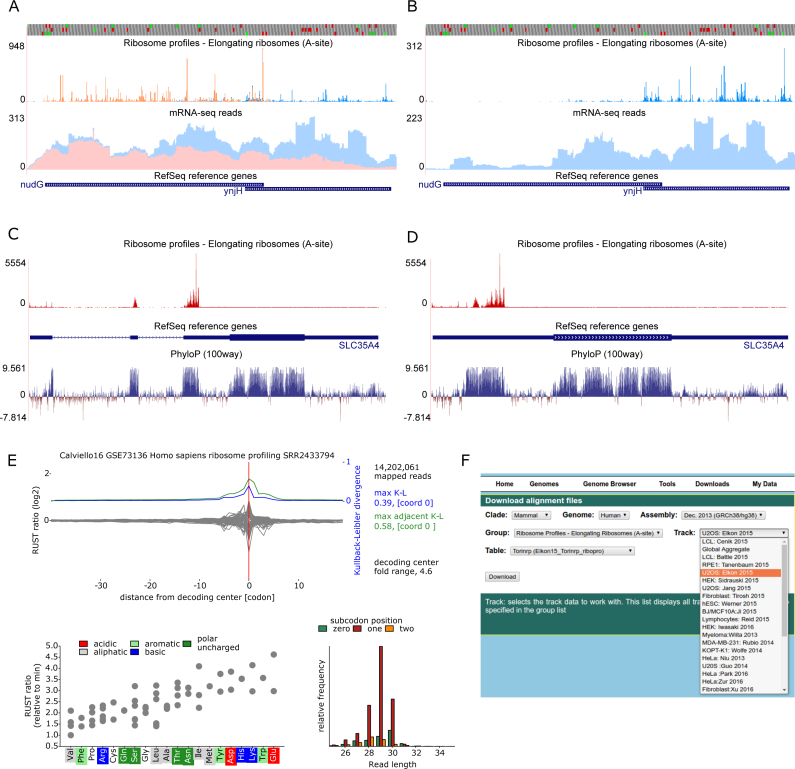
In addition to individual tracks reflecting Ribo-seq data generated under different conditions for each study, we aggregate each study's data into an All track. We then aggregate the All tracks from each study into a Global Aggregate track for each genome (Figure 1A–D). This has the effect of improving the overall Ribo-seq signal by reducing the contribution of dataset specific biases and stochastic noise due to low coverage. The increased number of datasets is expected to yield higher sensitivity. The Global Aggregate tracks are set as the default for each genome and users can turn on/off each study's data contribution to the aggregated data and then refine the visualizations by turning on/off individual tracks in each study. In addition, we provide Global Aggregate tracks through the UCSC Genome Browser for the human hg38 and hg19 assemblies.
Figure 1.
Exploring ribosome profiling data using GWIPS-viz. (A and B) Strand specific representation of the data for overlapping genes nudG and ynjH in the E. coli genome. In panel A, the Ribo-seq and mRNA-seq reads mapping to the forward strand (red) and to the reverse strand (blue) are both displayed. In panel B, only the reads mapping to the reverse strand are displayed. The profiles were generated using the Global aggregate tracks for E. coli in GWIPS-viz. (C and D) Aggregated human Ribo-seq data (red) at the SLC35A4 locus show that most of translation takes place at the uORF that spans the first three exons rather than the CDS (50,146,147). The exon-only view of the SLC35A4 locus improves the visualization of the translated uORF, the conservation of which is shown using the 100 vertebrates basewise conservation by PhyloP (148). (E) A RUST metafootprint profile that reveals the influence of mRNA codons on the relative read density in the vicinity of the ribosome is shown in grey in the top panel (145). The Kullback-Leibler divergence (blue for a single codon, green for adjacent codons) indicates the influence of each mRNA location on the frequency of ribosome footprint occurrence in the library. This is an example of a dataset with low sequencing biases, where the A-site codon influence is the highest. The lower left panel shows RUST estimates of relative codon decoding rates. The lower right panel shows the triplet periodicity signal (1,149) for individual read lengths. Panel E is taken from GWIPS-viz for study (20). (F) A screen-shot of the Downloads page that provides Ribo-seq and mRNA-seq read alignments for all tracks available in GWIPS-viz.
We have also incorporated a new type of data into GWIPS-viz. Recently the Preiss group developed a technique called translation complex profile sequencing (TCP-seq), where ribosome subunits are cross-linked to mRNA, allowing footprinting of both elongating and scanning ribosomes (15). We now provide this data in the Small Ribosomal Subunits (Footprints) track group for S. cerevisiae. Given that TCP-seq is a powerful tool for studying translation initiation (132), we anticipate that the experimental protocol will be adapted for other species quite soon and we will strive to incorporate new TCP-seq data into GWIPS-viz as it becomes available.
For S. cerevisiae we also generated an additional gene annotation track from transcript isoform sequencing (TIF-seq) data (133). Given that Saccharomyces Genome Database (SGD) (134) and Ensembl (135) gene annotations for S. cerevisiae do not include UTR regions, and given that Ribo-seq has shown that extensive translation occurs in 5′ leader regions (1) and to a lesser extent in 3′ trailer regions (97), we thought it useful to integrate the 5′ leader and 3′ trailer gene annotations as interpreted by (133) from their TIF-seq data for S. cerevisiae.
With respect to 5′ leader region delineation, we incorporated Riken 5′ cap analysis gene expression (CAGE) data (136) as a permanent track in the Annotations Tracks and External Data track group for the human hg19 assembly. At the time of the incorporation, Riken CAGE data tracks for other GWIPS-viz assemblies were not available for permanent track integration. However, as GWIPS-viz also now includes the UCSC Genome Browser's Track Hub functionality (137), we provide Riken's FANTOM5 tracks for hg38, hg19, mm9 and rn6 as public track hubs. While these tracks are hosted and managed by the Riken group on their own server, a simple connection makes it easy to explore their CAGE data in conjunction with our Ribo-seq/mRNA-seq tracks in GWIPS-viz.
Initially we did not provide UCSC Genome Browser's custom track feature (138) in GWIPS-viz. The custom track is only accessible to the user who uploads it, i.e. it is not a publicly available track. Many GWIPS-viz users, however, expressed an interest in the custom track feature as a means to explore their own Ribo-seq data in the context of published data and so we now include it. The custom track feature is also particularly useful for users of RiboGalaxy (139), a Galaxy based platform (140) that we have developed specifically for processing, mapping and analysing Ribo-seq data. Researchers can use the GWIPS-viz suite of tools in RiboGalaxy to generate Ribo-seq profiles that infer either the A-site (elongating ribosomes) or P-site (initiating ribosomes) from either the 5′ end or the 3′ end of Ribo-seq reads and the resulting profiles can be directly visualised as custom tracks in GWIPS-viz. The direct interface between GWIPS-viz and RiboGalaxy also allows data from GWIPS-viz to be retrieved into RiboGalaxy. We also provide a direct link to RiboGalaxy (http://ribogalaxy.ucc.ie/) from the GWIPS-viz homepage.
Improvements in data visualizations
Previously for bacterial genomes (E. coli K12, B. subtilis) our Ribo-seq and mRNA-seq profiles on GWIPS-viz did not provide strand orientation information. Our Ribo-seq density plots also used the center-weighted approach (141) to infer ribosome A-sites. Since then, several studies have shown that inferring the ribosome decoding center from the 3′ ends of bacterial Ribo-seq data is more accurate (124,142,143). We decided to carry-out an overhaul of our bacterial tracks and now provide strand orientation information using the UCSC Genome Browser overlay functionality (144) in addition to A-site inference using a fixed offset from 3′ footprint ends (Figure 1A,B). We have extended these improvements to the new bacterial genomes we now host in GWIPS-viz (E. coli BW25113, C. crescentus, S. coelicolor, S. aureus NCTC 8325, S. aureus USA300_FPR3757).
Recently we integrated the multi-region exon-only view (17), which is particularly useful for displaying Ribo-seq data for higher eukaryotes where exonic regions may be interrupted by long intronic regions (Figure 1C,D).
For higher eukaryotic datasets, we also now provide characteristics obtained with RUST (145). RUST utilizes Ribo-seq Unit Step Transfomation to normalize ribosome profiling data. It further provides characteristics of ribosome profiling datasets among which is a metafootprint profile which shows the difference between observed (experimental) and expected (equiprobable) frequencies of specific sequences (commonly codons) in the vicinity of a ribosome footprint. The expectation is that the highest variation in codon frequencies should occur at the ribosome decoding center (A-site) (Figure 1E). A high variation at the end of footprints would occur due to sequencing biases. Thus, metafootprint profiles can be used for assessing the level of sequencing biases in individual datasets. Clicking on each study link in the GWIPS-viz genome page will open a new page with the link to the RUST quality plots which include the RUST metafootprint profile as well as a plot showing triplet periodicity for reads of different lengths. The RUST plots also include a panel that shows the relative inferred A-site dwell times for each amino acid.
Downloading Ribo-seq and mRNA-seq alignments
Following user requests, we added the functionality to download our genomic alignments in bigWig format. While the Table Browser provides the option to download our Ribo-seq and mRNA-seq alignments in bedGraph format, many users requested our original alignment files. Hence, we built a separate Downloads page (Figure 1F) for this purpose. For each Ribo-seq study hosted on GWIPS-viz, users can download (1) ribosome profiles of elongating ribosomes (number of footprints whose inferred A-site match a specific coordinate, (2) Ribo-seq and (3) mRNA-seq coverage plots that provide the number of reads that map to each coordinate. Where available, data enriched with footprints of initiating ribosomes, represented as coordinates of inferred P-site codons, can also be downloaded. In addition, footprints of small ribosome subunits generated by TCP-seq (15) are available for download for S. cerevisiae as coverage plots.
FUTURE PLANS
The development of an automated computational data integration pipeline has greatly helped us to keep pace with the flux of new Ribo-seq data for genomes already existing in GWIPS-viz. We do, however, still have some backlog in terms of Ribo-seq data generated for genomes that we need to manually add to GWIPS-viz. We are examining ways in how we can improve our capacity to add new genomes particularly genomes that are not hosted on the UCSC Genome Browser.
We also aim to continue to improve the visualizations of Ribo-seq data. We wish to extend the overlay functionality with strand-specific display to all the data tracks in GWIPS-viz (currently provided for bacterial genomes only). Conversely, we want to provide RUST characteristics for Ribo-seq data generated for bacteria. This requires adapting the RUST programming code to using 3′ end offsetting for A-site inference which we plan to do soon. In addition, we want to use RUST parameters and other Ribo-seq specific parameters such as the triplet periodicity, to develop a quality scoring method and provide this information on GWIPS-viz for each dataset. This will allow users to compare Ribo-seq datasets in terms of their quality across studies. It will also help us and our user-base to determine what governs Ribo-seq data quality and how it can be maintained and improved in future Ribo-seq data.
We also wish to avail of new functionality as it becomes available on the UCSC Genome Browser. For this reason, we have carried out several upgrades of the GWIPS-viz browser to keep in-line with the UCSC Genome Browser. The recent exon-only view is one such example which is particularly beneficial for eukaryotic organisms that extensively use RNA splicing, permitting genomic alignments of Ribo-seq data to be explored without intervening introns.
DATA AVAILABILITY
GWIPS-viz is publicly and freely available at (http://gwips.ucc.ie/).
ACKNOWLEDGEMENTS
We wish to thank the following people who have helped at different stages with the continuous development of GWIPS-viz: Vimalkumar Velayudhan, Claire A Donohue, Svetlana Koroteeva, Kevin O’Riordan, Matthias Claes and Sandor Dedeyne. We also wish to thank the UCSC Genome Browser Development team who have always been so generous with their help.
Finally, we wish to thank all the world-wide users of GWIPS-viz as well as those users who have posted questions and suggestions on our GWIPS-viz forum (http://gwips.ucc.ie/Forum).
FUNDING
Science Foundation Ireland [12/IA/1335 to P.V.B]; A.M.M. and S.J.K. wish to acknowledge personal support from the Irish Research Council. Funding for open access charge: Science Foundation Ireland.
Conflict of interest statement. A.M.M. and P.V. B. are co-founders of Ribomaps Ltd., a company that offers ribosome profiling analysis that can be affected financially by the growing popularity of ribosome profiling.
REFERENCES
- 1. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weiss R.B., Atkins J.F.. Molecular biology. Translation goes global. Science. 2011; 334:1509–1510. [DOI] [PubMed] [Google Scholar]
- 3. Andreev D.E., O’Connor P.B., Loughran G., Dmitriev S.E., Baranov P.V., Shatsky I.N.. Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Res. 2017; 45:513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aramayo R., Polymenis M.. Ribosome profiling the cell cycle: lessons and challenges. Curr. Genet. 2017; doi:10.1007/s00294-017-0698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartholomaus A., Del Campo C., Ignatova Z.. Mapping the non-standardized biases of ribosome profiling. Biol. Chem. 2016; 397:23–35. [DOI] [PubMed] [Google Scholar]
- 6. Baudin-Baillieu A., Hatin I., Legendre R., Namy O.. Translation analysis at the genome scale by ribosome profiling. Methods Mol. Biol. 2016; 1361:105–124. [DOI] [PubMed] [Google Scholar]
- 7. Brar G.A., Weissman J.S.. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell. Biol. 2015; 16:651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gobet C., Naef F.. Ribosome profiling and dynamic regulation of translation in mammals. Curr. Opin. Genet. Dev. 2017; 43:120–127. [DOI] [PubMed] [Google Scholar]
- 9. Ingolia N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014; 15:205–213. [DOI] [PubMed] [Google Scholar]
- 10. Michel A.M., Baranov P.V.. Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale. Wiley Interdiscip. Rev. RNA. 2013; 4:473–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mumtaz M.A., Couso J.P.. Ribosomal profiling adds new coding sequences to the proteome. Biochem. Soc. Trans. 2015; 43:1271–1276. [DOI] [PubMed] [Google Scholar]
- 12. Parsons M., Myler P.J.. Illuminating parasite protein production by ribosome profiling. Trends Parasitol. 2016; 32:446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stern-Ginossar N. Decoding viral infection by ribosome profiling. J. Virol. 2015; 89:6164–6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stern-Ginossar N., Ingolia N.T.. Ribosome profiling as a tool to decipher viral complexity. Annu. Rev. Virol. 2015; 2:335–349. [DOI] [PubMed] [Google Scholar]
- 15. Archer S.K., Shirokikh N.E., Beilharz T.H., Preiss T.. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016; 535:570–574. [DOI] [PubMed] [Google Scholar]
- 16. Kodama Y., Shumway M., Leinonen R. International Nucleotide Sequence Database, C. . The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tyner C., Barber G.P., Casper J., Clawson H., Diekhans M., Eisenhart C., Fischer C.M., Gibson D., Gonzalez J.N., Guruvadoo L. et al. . The UCSC Genome Browser database: 2017 update. Nucleic Acids Res. 2017; 45:D626–D634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Michel A.M., Fox G., A M.K., De Bo C., O’Connor P.B., Heaphy S.M., Mullan J.P., Donohue C.A., Higgins D.G., Baranov P.V.. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 2014; 42:D859–D864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Battle A., Khan Z., Wang S.H., Mitrano A., Ford M.J., Pritchard J.K., Gilad Y.. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015; 347:664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calviello L., Mukherjee N., Wyler E., Zauber H., Hirsekorn A., Selbach M., Landthaler M., Obermayer B., Ohler U.. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods. 2016; 13:165–170. [DOI] [PubMed] [Google Scholar]
- 21. Cenik C., Cenik E.S., Byeon G.W., Grubert F., Candille S.I., Spacek D., Alsallakh B., Tilgner H., Araya C.L., Tang H. et al. . Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res. 2015; 25:1610–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Crappe J., Ndah E., Koch A., Steyaert S., Gawron D., De Keulenaer S., De Meester E., De Meyer T., Van Criekinge W., Van Damme P. et al. . PROTEOFORMER: deep proteome coverage through ribosome profiling and MS integration. Nucleic Acids Res. 2015; 43:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elkon R., Loayza-Puch F., Korkmaz G., Lopes R., van Breugel P.C., Bleijerveld O.B., Altelaar A.F., Wolf E., Lorenzin F., Eilers M. et al. . Myc coordinates transcription and translation to enhance transformation and suppress invasiveness. EMBO Rep. 2015; 16:1723–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gawron D., Ndah E., Gevaert K., Van Damme P.. Positional proteomics reveals differences in N-terminal proteoform stability. Mol. Syst. Biol. 2016; 12:858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo J.U., Agarwal V., Guo H., Bartel D.P.. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014; 15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012; 7:1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iwasaki S., Floor S.N., Ingolia N.T.. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016; 534:558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jakobsson M.E., Malecki J., Nilges B.S., Moen A., Leidel S.A., Falnes P.O.. Methylation of human eukaryotic elongation factor alpha (eEF1A) by a member of a novel protein lysine methyltransferase family modulates mRNA translation. Nucleic Acids Res. 2017; 45:8239–8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jang C., Lahens N.F., Hogenesch J.B., Sehgal A.. Ribosome profiling reveals an important role for translational control in circadian gene expression. Genome Res. 2015; 25:1836–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ji Z., Song R., Regev A., Struhl K.. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015; 4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Malecki J., Aileni V.K., Ho A.Y.Y., Schwarz J., Moen A., Sorensen V., Nilges B.S., Jakobsson M.E., Leidel S.A., Falnes P.O.. The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A). Nucleic Acids Res. 2017; 45:4370–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mills E.W., Wangen J., Green R., Ingolia N.T.. Dynamic regulation of a ribosome rescue pathway in erythroid cells and platelets. Cell Rep. 2016; 17:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niu Y., Zhao X., Wu Y.S., Li M.M., Wang X.J., Yang Y.G.. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013; 11:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park J.E., Yi H., Kim Y., Chang H., Kim V.N.. Regulation of Poly(A) tail and translation during the somatic cell cycle. Mol. Cell. 2016; 62:462–471. [DOI] [PubMed] [Google Scholar]
- 35. Raj A., Wang S.H., Shim H., Harpak A., Li Y.I., Engelmann B., Stephens M., Gilad Y., Pritchard J.K.. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. Elife. 2016; 5:e13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reid D.W., Shenolikar S., Nicchitta C.V.. Simple and inexpensive ribosome profiling analysis of mRNA translation. Methods. 2015; 91:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rubio C.A., Weisburd B., Holderfield M., Arias C., Fang E., DeRisi J.L., Fanidi A.. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014; 15:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C.. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sidrauski C., McGeachy A.M., Ingolia N.T., Walter P.. The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. Elife. 2015; 4:e05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Su X., Yu Y., Zhong Y., Giannopoulou E.G., Hu X., Liu H., Cross J.R., Ratsch G., Rice C.M., Ivashkiv L.B.. Interferon-gamma regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 2015; 16:838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tanenbaum M.E., Stern-Ginossar N., Weissman J.S., Vale R.D.. Regulation of mRNA translation during mitosis. Elife. 2015; 4:e07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tirosh O., Cohen Y., Shitrit A., Shani O., Le-Trilling V.T., Trilling M., Friedlander G., Tanenbaum M., Stern-Ginossar N.. The transcription and translation landscapes during human cytomegalovirus infection reveal novel host-pathogen interactions. PLoS Pathog. 2015; 11:e1005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wein N., Vulin A., Falzarano M.S., Szigyarto C.A., Maiti B., Findlay A., Heller K.N., Uhlen M., Bakthavachalu B., Messina S. et al. . Translation from a DMD exon 5 IRES results in a functional dystrophin isoform that attenuates dystrophinopathy in humans and mice. Nat. Med. 2014; 20:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Werner A., Iwasaki S., McGourty C.A., Medina-Ruiz S., Teerikorpi N., Fedrigo I., Ingolia N.T., Rape M.. Cell-fate determination by ubiquitin-dependent regulation of translation. Nature. 2015; 525:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wiita A.P., Ziv E., Wiita P.J., Urisman A., Julien O., Burlingame A.L., Weissman J.S., Wells J.A.. Global cellular response to chemotherapy-induced apoptosis. Elife. 2013; 2:e01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wolfe A.L., Singh K., Zhong Y., Drewe P., Rajasekhar V.K., Sanghvi V.R., Mavrakis K.J., Jiang M., Roderick J.E., Van der Meulen J. et al. . RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014; 513:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu B., Gogol M., Gaudenz K., Gerton J.L.. Improved transcription and translation with L-leucine stimulation of mTORC1 in Roberts syndrome. BMC Genomics. 2016; 17:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yoon J.H., De S., Srikantan S., Abdelmohsen K., Grammatikakis I., Kim J., Kim K.M., Noh J.H., White E.J., Martindale J.L. et al. . PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 2014; 5:5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zur H., Aviner R., Tuller T.. Complementary post transcriptional regulatory information is detected by PUNCH-P and ribosome profiling. Sci. Rep. 2016; 6:21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Andreev D.E., O’Connor P.B., Fahey C., Kenny E.M., Terenin I.M., Dmitriev S.E., Cormican P., Morris D.W., Shatsky I.N., Baranov P.V.. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife. 2015; 4:e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao X., Wan J., Liu B., Ma M., Shen B., Qian S.B.. Quantitative profiling of initiating ribosomes in vivo. Nat. Methods. 2015; 12:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gonzalez C., Sims J.S., Hornstein N., Mela A., Garcia F., Lei L., Gass D.A., Amendolara B., Bruce J.N., Canoll P. et al. . Ribosome profiling reveals a cell-type-specific translational landscape in brain tumors. J. Neurosci. 2014; 34:10924–10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rooijers K., Loayza-Puch F., Nijtmans L.G., Agami R.. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat. Commun. 2013; 4:2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stumpf C.R., Moreno M.V., Olshen A.B., Taylor B.S., Ruggero D.. The translational landscape of the mammalian cell cycle. Mol. Cell. 2013; 52:574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Alvarez-Dominguez J.R., Zhang X., Hu W.. Widespread and dynamic translational control of red blood cell development. Blood. 2017; 129:619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Atger F., Gobet C., Marquis J., Martin E., Wang J., Weger B., Lefebvre G., Descombes P., Naef F., Gachon F.. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E6579–E6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Blanco S., Bandiera R., Popis M., Hussain S., Lombard P., Aleksic J., Sajini A., Tanna H., Cortes-Garrido R., Gkatza N. et al. . Stem cell function and stress response are controlled by protein synthesis. Nature. 2016; 534:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Castaneda J., Genzor P., van der Heijden G.W., Sarkeshik A., Yates J.R. 3rd, Ingolia N.T., Bortvin A.. Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. EMBO J. 2014; 33:1999–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cho J., Yu N.K., Choi J.H., Sim S.E., Kang S.J., Kwak C., Lee S.W., Kim J.I., Choi D.I., Kim V.N. et al. . Multiple repressive mechanisms in the hippocampus during memory formation. Science. 2015; 350:82–87. [DOI] [PubMed] [Google Scholar]
- 60. de Klerk E., Fokkema I.F., Thiadens K.A., Goeman J.J., Palmblad M., den Dunnen J.T., von Lindern M., t Hoen P.A.. Assessing the translational landscape of myogenic differentiation by ribosome profiling. Nucleic Acids Res. 2015; 43:4408–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eichhorn S.W., Guo H., McGeary S.E., Rodriguez-Mias R.A., Shin C., Baek D., Hsu S.H., Ghoshal K., Villen J., Bartel D.P.. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell. 2014; 56:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fields A.P., Rodriguez E.H., Jovanovic M., Stern-Ginossar N., Haas B.J., Mertins P., Raychowdhury R., Hacohen N., Carr S.A., Ingolia N.T. et al. . A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol. Cell. 2015; 60:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fradejas-Villar N., Seeher S., Anderson C.B., Doengi M., Carlson B.A., Hatfield D.L., Schweizer U., Howard M.T.. The RNA-binding protein Secisbp2 differentially modulates UGA codon reassignment and RNA decay. Nucleic Acids Res. 2017; 45:4094–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gerashchenko M.V., Gladyshev V.N.. Ribonuclease selection for ribosome profiling. Nucleic Acids Res. 2017; 45:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hornstein N., Torres D., Das Sharma S., Tang G., Canoll P., Sims P.A.. Ligation-free ribosome profiling of cell type-specific translation in the brain. Genome Biol. 2016; 17:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Howard M.T., Carlson B.A., Anderson C.B., Hatfield D.L.. Translational redefinition of UGA codons is regulated by selenium availability. J. Biol. Chem. 2013; 288:19401–19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hurt J.A., Robertson A.D., Burge C.B.. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013; 23:1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J., Jackson S.E., Wills M.R., Weissman J.S.. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014; 8:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Janich P., Arpat A.B., Castelo-Szekely V., Lopes M., Gatfield D.. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 2015; 25:1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Laguesse S., Creppe C., Nedialkova D.D., Prevot P.P., Borgs L., Huysseune S., Franco B., Duysens G., Krusy N., Lee G. et al. . A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell. 2015; 35:553–567. [DOI] [PubMed] [Google Scholar]
- 71. Neri F., Rapelli S., Krepelova A., Incarnato D., Parlato C., Basile G., Maldotti M., Anselmi F., Oliviero S.. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017; 543:72–77. [DOI] [PubMed] [Google Scholar]
- 72. Reid D.W., Chen Q., Tay A.S., Shenolikar S., Nicchitta C.V.. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell. 2014; 158:1362–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reid D.W., Tay A.S., Sundaram J.R., Lee I.C., Chen Q., George S.E., Nicchitta C.V., Shenolikar S.. Complementary roles of GADD34- and CReP-containing eukaryotic initiation factor 2alpha phosphatases during the unfolded protein response. Mol. Cell. Biol. 2016; 36:1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sendoel A., Dunn J.G., Rodriguez E.H., Naik S., Gomez N.C., Hurwitz B., Levorse J., Dill B.D., Schramek D., Molina H. et al. . Translation from unconventional 5′ start sites drives tumour initiation. Nature. 2017; 541:494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. You K.T., Park J., Kim V.N.. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev. 2015; 29:2004–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Andreev D.E., O’Connor P.B., Zhdanov A.V., Dmitriev R.I., Shatsky I.N., Papkovsky D.B., Baranov P.V.. Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome Biol. 2015; 16:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ori A., Toyama B.H., Harris M.S., Bock T., Iskar M., Bork P., Ingolia N.T., Hetzer M.W., Beck M.. Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst. 2015; 1:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schafer S., Adami E., Heinig M., Rodrigues K.E., Kreuchwig F., Silhavy J., van Heesch S., Simaite D., Rajewsky N., Cuppen E. et al. . Translational regulation shapes the molecular landscape of complex disease phenotypes. Nat. Commun. 2015; 6:7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bazzini A.A., Johnstone T.G., Christiano R., Mackowiak S.D., Obermayer B., Fleming E.S., Vejnar C.E., Lee M.T., Rajewsky N., Walther T.C. et al. . Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014; 33:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chew G.L., Pauli A., Rinn J.L., Regev A., Schier A.F., Valen E.. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development. 2013; 140:2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee M.T., Bonneau A.R., Takacs C.M., Bazzini A.A., DiVito K.R., Fleming E.S., Giraldez A.J.. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013; 503:360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Subtelny A.O., Eichhorn S.W., Chen G.R., Sive H., Bartel D.P.. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014; 508:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Arnold A., Rahman M.M., Lee M.C., Muehlhaeusser S., Katic I., Gaidatzis D., Hess D., Scheckel C., Wright J.E., Stetak A. et al. . Functional characterization of C. elegans Y-box-binding proteins reveals tissue-specific functions and a critical role in the formation of polysomes. Nucleic Acids Res. 2014; 42:13353–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hendriks G.J., Gaidatzis D., Aeschimann F., Grosshans H.. Extensive oscillatory gene expression during C. elegans larval development. Mol. Cell. 2014; 53:380–392. [DOI] [PubMed] [Google Scholar]
- 85. Nedialkova D.D., Leidel S.A.. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015; 161:1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Aspden J.L., Eyre-Walker Y.C., Phillips R.J., Amin U., Mumtaz M.A., Brocard M., Couso J.P.. Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. Elife. 2014; 3:e03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kronja I., Yuan B., Eichhorn S.W., Dzeyk K., Krijgsveld J., Bartel D.P., Orr-Weaver T.L.. Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep. 2014; 7:1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Vasquez J.J., Hon C.C., Vanselow J.T., Schlosser A., Siegel T.N.. Comparative ribosome profiling reveals extensive translational complexity in different Trypanosoma brucei life cycle stages. Nucleic Acids Res. 2014; 42:3623–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Caro F., Ahyong V., Betegon M., DeRisi J.L.. Genome-wide regulatory dynamics of translation in the Plasmodium falciparum asexual blood stages. Elife. 2014; 3:e04106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Albert F.W., Muzzey D., Weissman J.S., Kruglyak L.. Genetic influences on translation in yeast. PLoS Genet. 2014; 10:e1004692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Baudin-Baillieu A., Legendre R., Kuchly C., Hatin I., Demais S., Mestdagh C., Gautheret D., Namy O.. Genome-wide translational changes induced by the prion [PSI+]. Cell Rep. 2014; 8:439–448. [DOI] [PubMed] [Google Scholar]
- 93. Beaupere C., Wasko B.M., Lorusso J., Kennedy B.K., Kaeberlein M., Labunskyy V.M.. CAN1 arginine permease deficiency extends yeast replicative lifespan via translational activation of stress response genes. Cell Rep. 2017; 18:1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cai Y., Futcher B.. Effects of the yeast RNA-binding protein Whi3 on the half-life and abundance of CLN3 mRNA and other targets. PLoS One. 2013; 8:e84630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dhungel N., Eleuteri S., Li L.B., Kramer N.J., Chartron J.W., Spencer B., Kosberg K., Fields J.A., Stafa K., Adame A. et al. . Parkinson's disease genes VPS35 and EIF4G1 interact genetically and converge on alpha-synuclein. Neuron. 2015; 85:76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gerashchenko M.V., Gladyshev V.N.. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014; 42:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Guydosh N.R., Green R.. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014; 156:950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jungfleisch J., Nedialkova D.D., Dotu I., Sloan K.E., Martinez-Bosch N., Bruning L., Raineri E., Navarro P., Bohnsack M.T., Leidel S.A. et al. . A novel translational control mechanism involving RNA structures within coding sequences. Genome Res. 2017; 27:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lareau L.F., Hite D.H., Hogan G.J., Brown P.O.. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014; 3:e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nissley D.A., Sharma A.K., Ahmed N., Friedrich U.A., Kramer G., Bukau B., O’Brien E.P.. Accurate prediction of cellular co-translational folding indicates proteins can switch from post- to co-translational folding. Nat. Commun. 2016; 7:10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Pop C., Rouskin S., Ingolia N.T., Han L., Phizicky E.M., Weissman J.S., Koller D.. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014; 10:770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schmidt C., Kowalinski E., Shanmuganathan V., Defenouillere Q., Braunger K., Heuer A., Pech M., Namane A., Berninghausen O., Fromont-Racine M. et al. . The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex. Science. 2016; 354:1431–1433. [DOI] [PubMed] [Google Scholar]
- 103. Sen N.D., Zhou F., Harris M.S., Ingolia N.T., Hinnebusch A.G.. eIF4B stimulates translation of long mRNAs with structured 5′ UTRs and low closed-loop potential but weak dependence on eIF4G. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:10464–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sen N.D., Zhou F., Ingolia N.T., Hinnebusch A.G.. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res. 2015; 25:1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Thiaville P.C., Legendre R., Rojas-Benitez D., Baudin-Baillieu A., Hatin I., Chalancon G., Glavic A., Namy O., de Crecy-Lagard V.. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell. 2016; 3:29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yerlikaya S., Meusburger M., Kumari R., Huber A., Anrather D., Costanzo M., Boone C., Ammerer G., Baranov P.V., Loewith R.. TORC1 and TORC2 work together to regulate ribosomal protein S6 phosphorylation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2016; 27:397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Young D.J., Guydosh N.R., Zhang F., Hinnebusch A.G., Green R.. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell. 2015; 162:872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zid B.M., O'Shea E.K.. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 2014; 514:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zinshteyn B., Gilbert W.V.. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013; 9:e1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Duncan C.D., Mata J.. The translational landscape of fission-yeast meiosis and sporulation. Nat. Struct. Mol. Biol. 2014; 21:641–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yu C.H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M.S., Liu Y.. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell. 2015; 59:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hsu P.Y., Calviello L., Wu H.L., Li F.W., Rothfels C.J., Ohler U., Benfey P.N.. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E7126–E7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Liu M.J., Wu S.H., Wu J.F., Lin W.D., Wu Y.C., Tsai T.Y., Tsai H.L., Wu S.H.. Translational landscape of photomorphogenic Arabidopsis. Plant Cell. 2013; 25:3699–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lukoszek R., Feist P., Ignatova Z.. Insights into the adaptive response of Arabidopsis thaliana to prolonged thermal stress by ribosomal profiling and RNA-Seq. BMC Plant Biol. 2016; 16:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lei L., Shi J., Chen J., Zhang M., Sun S., Xie S., Li X., Zeng B., Peng L., Hauck A. et al. . Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 2015; 84:1206–1218. [DOI] [PubMed] [Google Scholar]
- 116. Bartholomaus A., Fedyunin I., Feist P., Sin C., Zhang G., Valleriani A., Ignatova Z.. Bacteria differently regulate mRNA abundance to specifically respond to various stresses. Philos. Trans. A Math. Phys. Eng. Sci. 2016; 374:20150069. [DOI] [PubMed] [Google Scholar]
- 117. Guo M.S., Updegrove T.B., Gogol E.B., Shabalina S.A., Gross C.A., Storz G.. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014; 28:1620–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Haft R.J., Keating D.H., Schwaegler T., Schwalbach M.S., Vinokur J., Tremaine M., Peters J.M., Kotlajich M.V., Pohlmann E.L., Ong I.M. et al. . Correcting direct effects of ethanol on translation and transcription machinery confers ethanol tolerance in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2576–E2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Hwang J.Y., Buskirk A.R.. A ribosome profiling study of mRNA cleavage by the endonuclease RelE. Nucleic Acids Res. 2017; 45:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Latif H., Szubin R., Tan J., Brunk E., Lechner A., Zengler K., Palsson B.O.. A streamlined ribosome profiling protocol for the characterization of microorganisms. Biotechniques. 2015; 58:329–332. [DOI] [PubMed] [Google Scholar]
- 121. Li G.W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mohammad F., Woolstenhulme C.J., Green R., Buskirk A.R.. Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 2016; 14:686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Subramaniam A.R., Zid B.M., O'Shea E.K.. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell. 2014; 159:1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Woolstenhulme C.J., Guydosh N.R., Green R., Buskirk A.R.. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 2015; 11:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kannan K., Kanabar P., Schryer D., Florin T., Oh E., Bahroos N., Tenson T., Weissman J.S., Mankin A.S.. The general mode of translation inhibition by macrolide antibiotics. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15958–15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Subramaniam A.R., Deloughery A., Bradshaw N., Chen Y., O'Shea E., Losick R., Chai Y.. A serine sensor for multicellularity in a bacterium. Elife. 2013; 2:e01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Schrader J.M., Zhou B., Li G.W., Lasker K., Childers W.S., Williams B., Long T., Crosson S., McAdams H.H., Weissman J.S. et al. . The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet. 2014; 10:e1004463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Jeong Y., Kim J.N., Kim M.W., Bucca G., Cho S., Yoon Y.J., Kim B.G., Roe J.H., Kim S.C., Smith C.P. et al. . The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2). Nat. Commun. 2016; 7:11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Basu A., Yap M.N.. Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res. 2016; 44:4881–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Davis A.R., Gohara D.W., Yap M.N.. Sequence selectivity of macrolide-induced translational attenuation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15379–15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Michel A.M., Ahern A.M., Donohue C.A., Baranov P.V.. GWIPS-viz as a tool for exploring ribosome profiling evidence supporting the synthesis of alternative proteoforms. Proteomics. 2015; 15:2410–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Baranov P.V., Loughran G.. Catch me if you can: trapping scanning ribosomes in their footsteps. Nat. Struct. Mol. Biol. 2016; 23:703–704. [DOI] [PubMed] [Google Scholar]
- 133. Pelechano V., Wei W., Steinmetz L.M.. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013; 497:127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R. et al. . Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012; 40:D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Aken B.L., Ayling S., Barrell D., Clarke L., Curwen V., Fairley S., Fernandez Banet J., Billis K., Garcia Giron C., Hourlier T. et al. . The Ensembl gene annotation system. Database (Oxford). 2016; 2016:baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. FANTOM Consortium and the RIKEN PMI and CLST (DGT) Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T. et al. . A promoter-level mammalian expression atlas. Nature. 2014; 507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M. et al. . The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014; 42:D764–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Fujita P.A., Rhead B., Zweig A.S., Hinrichs A.S., Karolchik D., Cline M.S., Goldman M., Barber G.P., Clawson H., Coelho A. et al. . The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011; 39:D876–D882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Michel A.M., Mullan J.P., Velayudhan V., O’Connor P.B., Donohue C.A., Baranov P.V.. RiboGalaxy: A browser based platform for the alignment, analysis and visualization of ribosome profiling data. RNA Biol. 2016; 13:316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Eberhard C. et al. . The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016; 44:W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Oh E., Becker A.H., Sandikci A., Huber D., Chaba R., Gloge F., Nichols R.J., Typas A., Gross C.A., Kramer G. et al. . Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011; 147:1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Martens A.T., Taylor J., Hilser V.J.. Ribosome A and P sites revealed by length analysis of ribosome profiling data. Nucleic Acids Res. 2015; 43:3680–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. O’Connor P.B., Li G.W., Weissman J.S., Atkins J.F., Baranov P.V.. rRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics. 2013; 29:1488–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M. et al. . The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015; 43:D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. O’Connor P.B., Andreev D.E., Baranov P.V.. Comparative survey of the relative impact of mRNA features on local ribosome profiling read density. Nat. Commun. 2016; 7:12915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. et al. . A draft map of the human proteome. Nature. 2014; 509:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Vanderperre B., Lucier J.F., Bissonnette C., Motard J., Tremblay G., Vanderperre S., Wisztorski M., Salzet M., Boisvert F.M., Roucou X.. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One. 2013; 8:e70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Michel A.M., Choudhury K.R., Firth A.E., Ingolia N.T., Atkins J.F., Baranov P.V.. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2012; 22:2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
GWIPS-viz is publicly and freely available at (http://gwips.ucc.ie/).