Abstract
NLSdb is a database collecting nuclear export signals (NES) and nuclear localization signals (NLS) along with experimentally annotated nuclear and non-nuclear proteins. NES and NLS are short sequence motifs related to protein transport out of and into the nucleus. The updated NLSdb now contains 2253 NLS and introduces 398 NES. The potential sets of novel NES and NLS have been generated by a simple ‘in silico mutagenesis’ protocol. We started with motifs annotated by experiments. In step 1, we increased specificity such that no known non-nuclear protein matched the refined motif. In step 2, we increased the sensitivity trying to match several different families with a motif. We then iterated over steps 1 and 2. The final set of 2253 NLS motifs matched 35% of 8421 experimentally verified nuclear proteins (up from 21% for the previous version) and none of 18 278 non-nuclear proteins. We updated the web interface providing multiple options to search protein sequences for NES and NLS motifs, and to evaluate your own signal sequences. NLSdb can be accessed via Rostlab services at: https://rostlab.org/services/nlsdb/
INTRODUCTION
Eukaryotic cells are characterized by the envelopment of DNA into a membranous compartment, the nucleus (name: Greek ευ (eu) = well and καρυoν (karyon) = core). Shuttle proteins known as karyopherins, namely importins and exportins, facilitate the transport of proteins into and out of the nucleus through nuclear pore complexes (1–4). To identify their cargo, these proteins use specific sequence motifs: so-called nuclear localization signals (NLS) for the import into the nucleus, and nuclear export signals (NES) for the transport out of the nucleus.
NLS motifs vary substantially in terms of length and features (5–7). However, almost all share a simple feature, namely short stretches of mostly basic amino acids with the consensus sequence K-K/R-X-K/R. When a single such NLS on the sequence leads to import into the nucleus, these motifs are referred to as monopartite, while bipartite NLS often have two monopartite signals separated by a variable linker of typically 9–12 amino acids (8,9). The linker regions can be longer and tripartite motifs exist, as well. Many other types of NLS exist, e.g. the Proline-Tyrosine NLS (PY-NLS), named after the PY group in its motif: R/K/H-X(2–5)-P-Y (10).
The classical NES motif contains three to four hydrophobic amino acids, often leucine, and was first identified in HIV-1 (11,12). Several solutions to describing the consensus sequence of NES have been proposed (13–15), but they did not suffice to identify new NES-containing proteins (16).
Public databases such as UniProtKB (17), NESbase (14) and ValidNESs (18) have been able to collect just a few hundred experimentally verified NES and NLS motifs. Machine-learning methods targeted filling the gap, e.g. SeqNLS (19), NucPred (20), NESsential (16) and NLStradamus (21). However, as for every good prediction method, they make mistakes: those designed to predict nuclear proteins might mislabel non-nuclear proteins (false positives), and NES or NLS prediction methods might misplace the signal or miss it altogether (false negatives).
Our group has been developing generic machine learning-based methods predicting localization for almost two decades (22–26). In contrast, we developed NLSdb (27) as a comprehensive and specific database collecting as many experimentally verified NLS in a single resource as possible. In contrast to related resources before and after this, we also considered it to be important to enrich the resource by simple in silico analyses. For instance, although good experiments use controls for annotations, they cannot access as comprehensive datasets for controls as we can in silico: many motifs published by experimentalists mapped to many non-nuclear proteins, i.e. they were not specific enough (28), and many others mapped only to very few related proteins, i.e. were too specific (28). Experts tried to manoeuvre optimally between these two extremes beginning from 114 NLS carefully collected from the literature. Those were complemented by 194 potential NLS generated by ‘in silico mutagenesis’, i.e. mutated and tested by a computational algorithm essentially through iterating over the following simple steps (27,28): if too specific: make more general by shrinking motif, if too unspecific: lengthen motif. All operations (shorten/lengthen) were guided by multiple sequence alignments. Fifteen years later, we have now completed a major update to NLSdb using a similar algorithm, although this time we gave more control to the computer. We have also added NES motifs to the database.
MATERIALS AND METHODS
Datasets: nuclear and non-nuclear proteins
We downloaded all proteins from the UniProtKB/Swiss-Prot database (release May 2017), which had manually curated annotations for their subcellular localization (UniProtKB evidence code ECO:0000269). To reduce unrealistic protein fragments, we removed all proteins shorter than 50 residues. We masked low-complexity regions with SEG (29) and removed all proteins that did not have at least one segment of 30 consecutive unmasked residues.
We split the dataset into two distinct sets of nuclear and non-nuclear proteins. Proteins with experimental annotations for both nuclear and non-nuclear subcellular localizations were considered nuclear proteins. Although we previously ignored less reliable annotations (e.g. by similarity), we made an exception: if a protein was annotated experimentally as non-nuclear and it was annotated as nuclear by one of those less-reliable annotations, we removed it from our set. Analyzing the remaining set, we still found pairs of nuclear/non-nuclear proteins that had over 80% pairwise sequence identity. Though, the presence or absence of a single NLS could easily explain why two proteins N and C have >80% PIDE; and because N has the NLS and C does not, only N is nuclear. However, such observations might also suggest mistakes in the annotations. For security, we simply removed all such proteins from the set of non-nuclear proteins (toward this end we applied CD-Hit (30)). Our final datasets contained 8421 nuclear and 18 278 non-nuclear proteins.
Finally, we used UniqueProt (31) (HVAL > 0) to divide the set of nuclear proteins into 801 clusters of similar sequences, hereafter referred to as protein families. Those 801 families were used in the last step of our in silico mutagenesis algorithm (below).
Lists of NLS and NES
Next, we extracted all proteins from UniProtKB/Swiss-Prot (release May 2017) with nuclear export and localization signals (NES and NLS). We kept only proteins with the following evidence codes for their signal annotations: (i) ECO:0000269 (manually curated information for which there is published experimental evidence); (ii) ECO:0000305 (manually curated information which has been inferred by a curator based on his/her scientific knowledge or on the scientific content of an article); (iii) ECO:0000250 (manually curated information which has been propagated from a related experimentally characterized protein); and (iv) ECO:0000255 (manual assertions for information which has been generated by the UniProtKB automatic annotation system or by various sequence analysis programs). This resulted in 529 unique NES and 2362 unique NLS. We enriched this initial set tapping into other online sources, namely through 262 NES from ValidNESs, 80 NES from NESbase and 122 NLS from SeqNLS. Our final datasets contained 788 unique NES and 2466 unique NLS.
Protocol for in silico mutagenesis
Starting with our sets of collected NES and NLS, we applied an in silico mutagenesis algorithm to generate new potential signals. We applied the same steps to both sets (NLS and NES, for simplicity referred as ‘signals’ in the following).
(i) To ensure a high specificity for nuclear proteins, we removed all signals from the set that matched any of the non-nuclear proteins or did not match any of the nuclear proteins. To reduce the amount of non-functional residues (i.e. linker regions not related to the NLS), we removed signals longer than 30 residues. Next, we generated all possible variations of the remaining signals that differed by a single amino acid from the original sequence. Thus, for each signal we had 20 times its length variants, including the original one.
(ii) We iteratively applied the following two steps until no new variants could be generated: (a) remove all signals matching non-nuclear proteins or not matching nuclear proteins; (b) generate all possible variants by deleting a single residue, i.e. shortening the signal by one.
(iii) We removed signals that deviated too substantially from the features known for generic signals. We kept only NLS that contained at least three positively charged residues (H, K, R), two if they also included a PY-NLS motif (R/K/H-X(2–5)-P-Y). At least one region (two for NLS of 20 or more residues) had to have an overall positive charge (i.e. not cancelled out by negative residues right next to the positive ones). NES were removed if they had fewer than three hydrophobic residues (A, F, I, L, M, V), or if <30% of the signal was hydrophobic.
(iv) To filter out potential signals that are too specific, we kept only those that matched proteins from at least two families (as defined above). We removed redundant signals, i.e. for any pair of signals S1 and S2, if S1 is a substring of S2, we kept only the shorter signal S1.
Note that our final signals by design reached 100% specificity on the set of proteins that we used for development (signals were removed if they match non-nuclear proteins). In order to keep specificity high, we deliberately avoided to combine motifs in regular expressions. For instance, experimentally observing the motifs RKHEL and LEHKR does not imply that all motifs matched by [RL][KE]H[EK][LR] constitute a valid NLS. In particular, LEHEL is unlikely to function as NLS.
Dataset of whole proteomes
We analyzed the ‘entire proteomes’ for the nine eukaryotic organisms that contributed most to our set of experimentally annotated nuclear proteins (91%, Table 1). These were: Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, Mus musculus, Oryza sativa, Rattus norvegicus, Schizosaccharomyces pombe and Saccharomyces cerevisiae. We downloaded the latest reference proteomes (release April 2017) from the European Bioinformatics Institute (EMBL-EBI) (32).
Table 1. Proteins with NES and NLS in nuclear protein dataset.
| Organism | Number of nuclear proteins in NLSdb | Proteins with NLS | Proteins with NES |
|---|---|---|---|
| Homo sapiens (human) | 2163 | 820 (37.9%) | 185 (8.6%) |
| Schizosaccharomyces pombe (fission yeast) | 1263 | 282 (22.3%) | 52 (4.1%) |
| Arabidopsis thaliana (thale cress) | 1241 | 430 (34.6%) | 37 (3.0%) |
| Mus musculus (mouse) | 1011 | 420 (41.5%) | 89 (8.8%) |
| Saccharomyces cerevisiae (brewer's yeast) | 1010 | 294 (29.1%) | 33 (3.3%) |
| Drosophila melanogaster (fruit fly) | 334 | 149 (44.6%) | 20 (6.0%) |
| Caenorhabditis elegans (roundworm) | 273 | 113 (41.4%) | 19 (7.0%) |
| Rattus norvegicus (rat) | 237 | 85 (35.9%) | 22 (9.3%) |
| Oryza sativa (rice) | 140 | 46 (32.9%) | 5 (3.6%) |
| Sum nine organisms | 7672 | 2639 (34.4%) | 462 (6.0%) |
Organism: latin (common) names for the nine organisms that contributed the most nuclear proteins (sorted by number of nuclear proteins) to NLSdb (together 7672 proteins in these nine organisms accounted for 91% of all currently known 8421 nuclear proteins); Number of nuclear proteins in NLSdb: gives the number of proteins annotated experimentally as nuclear and retained in NLSdb after applying a variety of filters (Methods); Proteins with NLS/NES: numbers and fractions (brackets) of the nuclear proteins that contain at least one NLS or NES from NLSdb.
In addition, we downloaded the subcellular localization annotations for 12 003 human proteins from the Human Protein Atlas (release October 2017) (33,34). However, we only used annotations classified as either ‘validated’ or ‘supported’, and ignored the less reliable ‘approved’ and ‘uncertain’.
RESULTS AND DISCUSSION
Database growth
Our in silico mutagenesis protocol generated 1177 potential NES and 5189 potential NLS from the initial collection of 788 original NES and 2466 original NLS. Of those, 192 potential NES and 1651 potential NLS matched at least two different protein families. In addition to those, NLSdb includes all original NES and NLS that match nuclear proteins, while not matching non-nuclear proteins. The final list is: 398 NES and 2253 NLS.
In comparison: the original NLSdb contained only 308 experimental and potential NLS, and no NES. Those 308 NLS match 1810 (21%) of the 8421 nuclear proteins, but also 1017 of the non-nuclear proteins. Considering only NLS motifs that exclusively match nuclear proteins, the previous version covered only 174 (2%) proteins. In contrast, the NLS of the new NLSdb cover 2928 (35%) of the nuclear proteins. In other words, the error-free coverage of the new version increased by a factor of 17 (factor of 1.6 if ignoring the errors). This also highlighted why we moved from regular expressions (old version) to lists of signal sequences (new version): regular expressions are prone to quickly becoming too permissive leading to matches in non-nuclear proteins.
Specificity and sensitivity of NES and NLS
Only 206 of the collected 788 NES (26%) matched at least one of the 8421 nuclear proteins without matching any of the 18 278 non-nuclear proteins. In fact, only 231 NES (29%) matched any experimentally annotated nuclear protein. Similarly, only 639 (26%) of the annotated 2466 NLS matched known nuclear proteins without matching non-nuclear proteins, and only 969 NLS (39%) matched any known nuclear protein. In fact, for most proteins with NES and NLS annotations an experimental annotation as nuclear proteins was missing. Only 17% (754 of 4429) of the proteins we used to extract NES and NLS from UniProtKB/Swiss-Prot were experimentally annotated as nuclear.
According to our protocol (Methods: In silico mutagenesis), we retained only the 206 NES and 639 NLS that exclusively matched known nuclear proteins. These motifs matched 191 (NES) and 713 (NLS) of the annotated 8421 nuclear proteins. This suggests that most motifs are too specific, i.e. too long to match any other sequence. While such specific motifs might be biologically meaningful, they are not very helpful to discover motifs in proteins outside of the well annotated families.
The in silico-refined signals in NLSdb were both more sensitive (i.e. matching more proteins) and more specific (i.e. matching only nuclear proteins): the 192 NES matched 439 nuclear proteins from 265 families, and the 1651 NLS in 2773 nuclear proteins from 618 families. Thereby the average number of nuclear proteins matched by one motif increased from 1.2 for the original NES to 2.6 for the potential NES and from 1.3 for the original NLS to 3.2 for the potential NLS.
The bias in the dominance of particular organisms to the final set appeared to mirror the bias of experimental biology with most annotations from human, relatively few from plants, and relatively many from yeast (Table 1). However, this bias largely disappeared when considering how many proteins of an organism the NLS/NES matched. For instance, although almost ten times more nuclear proteins from human were annotated than from rat, the NLS from NLSdb matched 38% of the experimentally annotated human and 36% of the experimentally annotated rat proteins, i.e. rather similar fractions of the annotated nuclear proteins. The same held for NES.
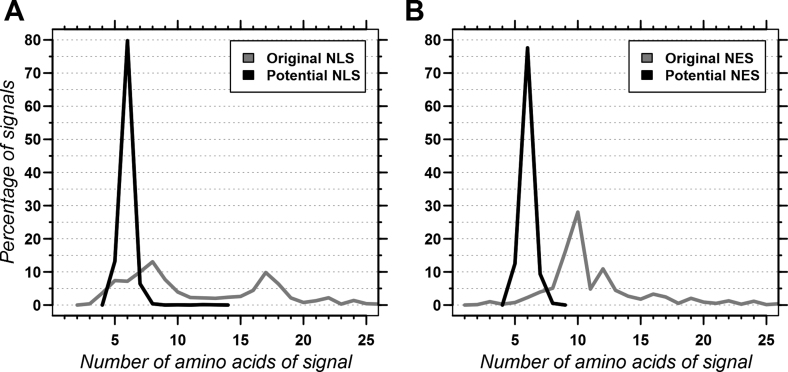
The potential NES and NLS in NLSdb were rather short (Figure 1) because our in silico mutagenesis protocol can only shorten NES and NLS. Furthermore, we kept only the shorter of two redundant motifs. Almost all of the resulting motifs (NLS and NES) were about six amino acids long. This implies that a match in a protein sequence indicates the site of a NES or NLS, but that the signal tagged by NLSdb might not suffice to facilitate nuclear transport on its own. Instead, the biologically relevant NLS/NES might require additional neighbouring residues to bind to shuttle proteins (karyopherins). For example, two consecutive matches in a protein might indicate a bipartite NLS, while the same signal might match a monopartite NLS in another protein.
Figure 1.
Length distribution of NLS and NES sequences. The graphs compare the length distribution for the original NLS (A: gray line; total 2466 NLS) and the NLSdb set of NLS refined through in silico mutagenesis (A: dark line; total 1651 NLS), as well as the corresponding distributions for the original NES (B: gray line; total 788) and the NLSdb refined set of NES (B: dark line; total 192 NES). Note that motifs with over 25 amino acids were observed, but are not shown in the graphs due to sparseness (total: 156 original NLS, 42 original NES).
NES and NLS in NLSdb map to about half of all nuclear proteins in popular organisms
The enriched dataset of motifs available through NLSdb matched many proteins in all nine organisms analyzed (Table 2). On average, NLS motifs matched 9–13% of all proteins in each proteome and NES motifs 1–2% (Table 2). Comparing those numbers to the estimated fraction of nuclear proteins predicted by LocTree3 (22), suggested a coverage very similar to that estimated by the set of experimentally verified nuclear proteins (Table 1 versus Table 2). For example, the NLS cover 38% of the experimentally annotated human nuclear proteins, and about 42% of the human nuclear proteins predicted by LocTree3. Overall, the fractions of predicted nuclear proteins suggested that about 30–40% of all nuclear proteins in those nine proteomes were matched by one of the NLS in NLSdb.
Table 2. Nuclear signals and proteins in entire proteomes.
| Organism | Number of proteins | Proteins with NLS | Proteins with NES | LocTree3 (nucleus) |
|---|---|---|---|---|
| Homo sapiens (human) | 21 042 | 2673 (12.7%) | 375 (1.8%) | 30% |
| Schizosaccharomyces pombe (fission yeast) | 5142 | 501 (9.7%) | 78 (1.5%) | 34% |
| Arabidopsis thaliana (thale cress) | 27 502 | 2768 (10.1%) | 246 (0.9%) | 31% |
| Mus musculus (mouse) | 22 262 | 2684 (12.1%) | 358 (1.6%) | 30% |
| Saccharomyces cerevisiae (brewer's yeast) | 6722 | 681 (10.1%) | 63 (0.9%) | 31% |
| Drosophila melanogaster (fruit fly) | 13 757 | 1573 (11.4%) | 168 (1.2%) | 31% |
| Caenorhabditis elegans (roundworm) | 20 057 | 1759 (8.8%) | 170 (0.8%) | 27% |
| Rattus norvegicus (rat) | 21 412 | 2555 (11.9%) | 359 (1.7%) | 28% |
| Oryza sativa (rice) | 44 321 | 4285 (9.7%) | 280 (0.6%) | 28% |
Organism: latin (common) names for the nine organisms that contributed the most nuclear proteins to NLSdb (sorted by number of nuclear proteins); Number of proteins: gives the number of proteins found in the ‘entire proteome’ as we accessed it (Methods: Dataset of whole proteomes); Proteins with NLS/NES: numbers and fractions (brackets) of the nuclear proteins that contain at least one NLS or NES from NLSdb; LocTree3 (nucleus): lists the fractions of proteins predicted by our generic machine learning-based method LocTree3 (22) as nuclear.
We tried to validate the specificity of our NLS and NES on the set of potential novel human nuclear proteins by comparing these proteins to annotations from the Human Protein Atlas. Of the 2673 human proteins containing NLS from NLSdb (Table 2), 1853 proteins were not in our dataset of experimentally annotated nuclear proteins from UniProtKB/Swiss-Prot, therefore representing potential novel nuclear proteins. For 516 of them we had subcellular localization annotations from the Human Protein Atlas. There, 407 (79%) of the 516 proteins were annotated as nuclear proteins, implying an error rate of 21% (i.e. proteins containing NLS being non-nuclear). Repeating the same process for the 375 human proteins containing NES (Table 2), we found 65% (34 of 52) of potential novel proteins to be annotated as nuclear. This unfortunately showed that despite our careful approach the signals were not always 100% specific. However, we would like to note that for all 2597 proteins, which were in both the Human Protein Atlas and our entire datasets of experimentally annotated nuclear and non-nuclear proteins, the agreement on being nuclear or non-nuclear was only 90%. Thus, the 79 and 65% should be compared to an upper limit of 90% and not to 100%.
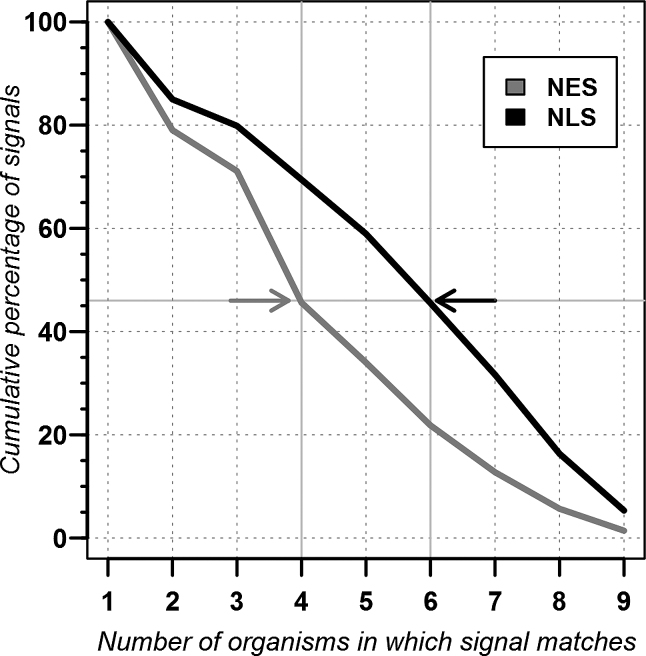
Our in silico mutagenesis protocol succeeded in rendering signals that are found in many organisms. For instance, 46% of the NES matched in at least four of nine model organisms, and 46% of the NLS matched in six or more (Figure 2 arrows and Table 1). Only about 1% of all NES and 5% of all NLS matched in all nine organisms.
Figure 2.
NES and NLS common to multiple organisms. The graph shows the cumulative percentage of NES and NLS found in at least one protein from the nine organisms used in Tables 1–2 (Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, Mus musculus, Oryza sativa, Rattus norvegicus, Schizosaccharomyces pombe and Saccharomyces cerevisiae). Hundred percent corresponds to 353 NES and 2180 NLS contained in NLSdb. For instance, 46% of the NES and 46% of the NLS matched in at least four and six organisms, respectively (arrows).
NLSdb web interface
In addition to updating the dataset of NES and NLS, we also re-designed the NLSdb web interface. It now supports a wider range of different query types as well as batch queries, i.e. submitting more than one query at a time. Queries can be directly inputted as text or uploaded as a file.
Searching proteins for NES and NLS
Users can submit protein sequences to find potential NES and NLS. The server accepts sequences in FASTA format or through their UniProtKB accession numbers. If any NES or NLS match the sequences, NLSdb reports the position and sequence of the hit as well as two confidence scores. These confidence scores are the number of nuclear proteins and families matched by the hit.
Searching and evaluating NES and NLS
Users can also submit their own putative NES or NLS signals and either compare those against our dataset or evaluate them on the set of nuclear and non-nuclear proteins. NLSdb returns all NES and NLS that are matched by the query. Submitting a signal for evaluation returns all nuclear and non-nuclear proteins the query is matching.
Browsing and downloading the database
NLSdb provides a browse function where users can simply look at all the NES and NLS sequences within NLSdb, as well as those collected from other databases. Each entry includes the following information: (i) type of the signal (possible values: NES or NLS); (ii) number of nuclear proteins and families matched (integers 0-N); (iii) type of evidence for annotation (possible values: experimental, determined by an expert or potential); (iv) database from which motif was extracted (possible values: UniProtKB AC, NESbase, ValidNESs, SeqNLS or in silico mutagenesis).
Users can also download all available data: the sets of collected and generated NES and NLS, and the sets of nuclear and non-nuclear proteins. This can be useful if a user wants to check several thousands of sequences on his local machine and does not need all the information provided by NLSdb, e.g. wants only to check if a NES or NLS is present. Currently all downloads are available as comma separated value files.
CONCLUSION
The substantially updated version of NLSdb makes available NLS and NES that appeared to match about 34 and 6%, respectively, of all the nuclear proteins in nine model organisms (A. thaliana, C. elegans, D. melanogaster, H. sapiens, M. musculus, O. sativa, R. norvegicus, S. pombe and S. cerevisiae). Before the update, the specific motifs contained in the previous version of NLSdb matched 17-times fewer proteins than the new version. When also allowing error-prone motifs (matching non-nuclear proteins), the old version covered 1.6-times fewer proteins than the new version. The new NLSdb contains about 7-times as many potential signals that help to understand the molecular mechanism of nuclear transport for many uncharacterized proteins. Along with a complete redesign of the interface, this overhauled resource might help in the design and follow up analysis of many experiments.
DATA AVAILABILITY
The complete sets of nuclear and non-nuclear proteins, as well as the sets of collected and potential NES and NLS, is available at the NLSdb website: https://rostlab.org/services/nlsdb/.
ACKNOWLEDGEMENTS
We thank Tim Karl for technical and Inga Weise (both TUM) for administrative assistance. Thanks to Kristoffer Rapacki and Søren Brunak (NESbase), Hsueh-Fen Juan (ValidNESs), Jianjun Hu (SeqNLS) and all the other people that worked on those datasets and made them easily available. Last, but not least, thanks to all who practice open science and deposit their data into public databases, and those who maintain these excellent databases.
FUNDING
Alexander von Humboldt Foundation through German Federal Ministry for Education and Research; Bavarian Competence Network for Technical and Scientific High Performance Computing (KONWIHR-III) (to M.B.); Ernst Ludwig Ehrlich Studienwerk (to T.G.). Funding for open access charge: German Research Foundation (DFG) and Technical University of Munich (TUM) in the framework of the Open Access Publishing Program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Freitas N., Cunha C.. Mechanisms and signals for the nuclear import of proteins. Curr. Genomics. 2009; 10:550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mosammaparast N., Pemberton L.F.. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004; 14:547–556. [DOI] [PubMed] [Google Scholar]
- 3. Fried H., Kutay U.. Nucleocytoplasmic transport: taking an inventory. Cell Mol. Life Sci. 2003; 60:1659–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003; 112:441–451. [DOI] [PubMed] [Google Scholar]
- 5. Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., Corbett A.H.. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2007; 282:5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hodel M.R., Corbett A.H., Hodel A.E.. Dissection of a nuclear localization signal. J. Biol. Chem. 2001; 276:1317–1325. [DOI] [PubMed] [Google Scholar]
- 7. Gorlich D. Nuclear protein import. Curr. Opin. Cell Biol. 1997; 9:412–419. [DOI] [PubMed] [Google Scholar]
- 8. Bickmore W.A., Sutherland H.G.. Addressing protein localization within the nucleus. EMBO J. 2002; 21:1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gorlich D., Kutay U.. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999; 15:607–660. [DOI] [PubMed] [Google Scholar]
- 10. Lee B.J., Cansizoglu A.E., Suel K.E., Louis T.H., Zhang Z., Chook Y.M.. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006; 126:543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wen W., Meinkoth J.L., Tsien R.Y., Taylor S.S.. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995; 82:463–473. [DOI] [PubMed] [Google Scholar]
- 12. Fischer U., Huber J., Boelens W.C., Mattaj I.W., Luhrmann R.. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995; 82:475–483. [DOI] [PubMed] [Google Scholar]
- 13. la Cour T., Kiemer L., Molgaard A., Gupta R., Skriver K., Brunak S.. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004; 17:527–536. [DOI] [PubMed] [Google Scholar]
- 14. la Cour T., Gupta R., Rapacki K., Skriver K., Poulsen F.M., Brunak S.. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003; 31:393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bogerd H.P., Fridell R.A., Benson R.E., Hua J., Cullen B.R.. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 1996; 16:4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fu S.C., Imai K., Horton P.. Prediction of leucine-rich nuclear export signal containing proteins with NESsential. Nucleic Acids Res. 2011; 39:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. The UniProt, C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu S.C., Huang H.C., Horton P., Juan H.F.. ValidNESs: a database of validated leucine-rich nuclear export signals. Nucleic Acids Res. 2013; 41:D338–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin J.R., Hu J.. SeqNLS: nuclear localization signal prediction based on frequent pattern mining and linear motif scoring. PLoS One. 2013; 8:e76864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brameier M., Krings A., MacCallum R.M.. NucPred–predicting nuclear localization of proteins. Bioinformatics. 2007; 23:1159–1160. [DOI] [PubMed] [Google Scholar]
- 21. Nguyen Ba A.N., Pogoutse A., Provart N., Moses A.M.. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009; 10:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goldberg T., Hecht M., Hamp T., Karl T., Yachdav G., Ahmed N., Altermann U., Angerer P., Ansorge S., Balasz K. et al. LocTree3 prediction of localization. Nucleic Acids Res. 2014; 42:W350–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andrade M.A., O’Donoghue S.I., Rost B.. Adaptation of protein surfaces to subcellular location. J. Mol. Biol. 1998; 276:517–525. [DOI] [PubMed] [Google Scholar]
- 24. Nair R., Rost B.. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 2005; 348:85–100. [DOI] [PubMed] [Google Scholar]
- 25. Nair R., Rost B.. Better prediction of sub-cellular localization by combining evolutionary and structural information. Proteins. 2003; 53:917–930. [DOI] [PubMed] [Google Scholar]
- 26. Goldberg T., Hamp T., Rost B.. LocTree2 predicts localization for all domains of life. Bioinformatics. 2012; 28:i458–i465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nair R., Carter P., Rost B.. NLSdb: database of nuclear localization signals. Nucleic Acids Res. 2003; 31:397–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cokol M., Nair R., Rost B.. Finding nuclear localisation signals. EMBO Rep. 2000; 1:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wootton J.C., Federhen S.. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996; 266:554–571. [DOI] [PubMed] [Google Scholar]
- 30. Li W., Godzik A.. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006; 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- 31. Mika S., Rost B.. UniqueProt: creating representative protein sequence sets. Nucleic Acids Res. 2003; 31:3789–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altenhoff A.M., Boeckmann B., Capella-Gutierrez S., Dalquen D.A., DeLuca T., Forslund K., Huerta-Cepas J., Linard B., Pereira C., Pryszcz L.P. et al. Standardized benchmarking in the quest for orthologs. Nat. Methods. 2016; 13:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L.M. et al. A subcellular map of the human proteome. Science. 2017; 356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 34. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S. et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010; 28:1248–1250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete sets of nuclear and non-nuclear proteins, as well as the sets of collected and potential NES and NLS, is available at the NLSdb website: https://rostlab.org/services/nlsdb/.