Abstract
GenBank® (www.ncbi.nlm.nih.gov/genbank/) is a comprehensive database that contains publicly available nucleotide sequences for 400 000 formally described species. These sequences are obtained primarily through submissions from individual laboratories and batch submissions from large-scale sequencing projects, including whole genome shotgun and environmental sampling projects. Most submissions are made using BankIt, the National Center for Biotechnology Information (NCBI) Submission Portal, or the tool tbl2asn. GenBank staff assign accession numbers upon data receipt. Daily data exchange with the European Nucleotide Archive and the DNA Data Bank of Japan ensures worldwide coverage. GenBank is accessible through the NCBI Nucleotide database, which links to related information such as taxonomy, genomes, protein sequences and structures, and biomedical journal literature in PubMed. BLAST provides sequence similarity searches of GenBank and other sequence databases. Complete bimonthly releases and daily updates of the GenBank database are available by FTP. Recent updates include changes to sequence identifiers, submission wizards for 16S and Influenza sequences, and an Identical Protein Groups resource.
INTRODUCTION
GenBank (1) is a comprehensive public database of nucleotide sequences and supporting bibliographic and biological annotations. GenBank is built and distributed by the National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine (NLM), located on the campus of the US National Institutes of Health (NIH) in Bethesda, MD, USA.
NCBI builds GenBank primarily from submissions of sequence data from authors and from bulk submissions of whole-genome shotgun (WGS) and other high-throughput data from sequencing centers. The US Patent and Trademark Office also contributes sequences from issued patents. GenBank participates with the EMBL-EBI European Nucleotide Archive (ENA) (2) and the DNA Data Bank of Japan (DDBJ) (3) as a partner in the International Nucleotide Sequence Database Collaboration (INSDC) (4). The INSDC partners exchange data daily to ensure that a uniform and comprehensive collection of sequence information is available worldwide. NCBI makes GenBank data available at no cost through the Internet, FTP and a wide range of web-based retrieval and analysis services (5).
RECENT DEVELOPMENTS
Changes to sequence identifiers
As first described in the release notes for GenBank 199.0 in December 2013, and discussed in more detail previously (1), NCBI is phasing out the practice of assigning GI numbers as sequence identifiers. As time progresses, we will no longer assign GI numbers to a gradually growing number of new sequences. (Current examples of such sequences are unannotated contigs in WGS and TSA projects.) In November 2016, we removed GI numbers from the default flat file presentations and FASTA definition lines of sequence data records, whether obtained from the web, API calls, or the NCBI FTP site. GenBank release 217 was the last release to contain GI numbers in the standard flat file distribution. Going forward, sequence records with existing GI numbers will retain them in XML and Abstract Syntax Notation One (ASN.1) formats, and NCBI services that accept GI numbers as input will continue to be supported. The preferred identifier for sequence records is now the accession.version. For example, the E-utilities now accept accession.version identifiers as input and can provide them as output when the parameter idtype is set to ‘acc’.
Ribosomal RNA submission wizard
The rRNA submission wizard, part of the NCBI submission portal, now offers faster, real-time analysis to assist submitters of rRNA sequences from both prokaryotes and eukaryotes (submit.ncbi.nlm.nih.gov/genbank/help/). Prokaryotic samples can be from uncultured, environmental sources, or pure cultured strains, and can include 16S rRNA, 23S rRNA, or 16S-23S rRNA intergenic spacers. Eukaryotic samples can include both large and small subunit rRNA, nuclear rRNA-ITS regions, and internal transcribed spacers. If samples were generated using next-generation technologies, only assembled sequences (two or more reads) will be accepted. Sequences submitted using the wizard will be automatically processed and checked for chimeras, vector contamination, low quality sequence, and other problems.
Batch genome submissions
The NCBI submission portal now supports the submission of up to 400 genomes in a single set. These genomes can be either prokaryotic or eukaryotic, and can either be WGS or non-WGS (but all sequences in the batch must be either WGS or non-WGS; mixed sets are not allowed). Viral and phage genomes are not currently accepted using this mechanism. Currently batch genome submissions have other requirements, including that all sequences in the batch belong to the same BioProject, that they have the same initial release date, and that each genome have a separate file. We are exploring the possibility of allowing batch submissions for multiple BioProjects. A complete list of requirements is available (www.ncbi.nlm.nih.gov/genbank/genomesubmit/).
Influenza submission wizard
NCBI has released a new wizard that supports the submission of Influenza sequences. The wizard accepts Influenza A, B, and C submissions, but only sequences from one viral type may be included in a single submission. In addition to validating the data, the wizard produces a standard strain identifier based on submitted metadata such as the isolate, place of collection, collection date, host, and serotype. NCBI will then annotate the submission using the influenza virus annotation tool (www.ncbi.nlm.nih.gov/genomes/FLU/annotation/), and results will be sent to the submitter, including any errors that need correcting.
Identical protein groups
In 2013 NCBI introduced non-redundant protein sequences (with accessions beginning with WP) that represent sets of identical proteins annotated on prokaryotic genomes (6). To clarify the relationships between these WP sequences and the set of individual Nucleotide CDS sequences they represent, in 2014 NCBI added the ‘Identical Protein Report’ to the Protein database. Now these reports have been improved and collected in a new resource called Identical Protein Groups (www.ncbi.nlm.nih.gov/ipg/). This resource includes all NCBI protein sequences, including records from INSDC, RefSeq, Swiss-Prot, and PDB, with links to nucleotide coding sequences from GenBank and RefSeq. The title of each record is derived from the ‘best’ sequence in each group, where the hierarchy for determining the best sequence is RefSeq > Swiss-Prot > PIR, PDB > GenBank > patent. Searches in this database can be filtered by database source, taxonomy, and the number of sequences in the group. These reports continue to be available through the E-utility EFetch with &db = protein&rettype = ipg (eutils.ncbi.nlm.nih.gov).
ORGANIZATION OF THE DATABASE
GenBank divisions
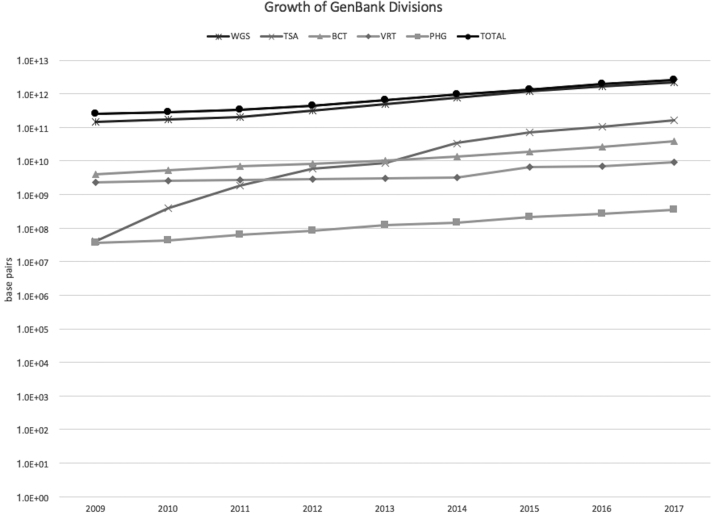
GenBank assigns sequence records to various divisions based either on the source taxonomy or the sequencing strategy used to obtain the data. There are twelve taxonomic divisions (BCT, ENV, INV, MAM, PHG, PLN, PRI, ROD, SYN, UNA, VRL, VRT) and five high-throughput divisions (EST, GSS, HTC, HTG, STS). In addition, the PAT division contains records supplied by patent offices, the TSA division contains sequences from transcriptome shotgun assembly (TSA) projects, and the WGS division contains sequences from whole genome shotgun projects. The size and growth of these divisions, and of GenBank as a whole, are shown in Table 1 and Figure 1.
Table 1. Growth of GenBank Divisions (nucleotide base-pairs).
| Division | Description | Release 221 (August 2017) | Annual increase (%)* |
|---|---|---|---|
| TSA | Transcriptome shotgun assembly | 167 045 663 417 | 61.55 |
| BCT | Bacteria | 39 102 455 601 | 47.70 |
| WGS | Whole genome shotgun data | 2 242 294 609 510 | 36.96 |
| VRT | Other vertebrates | 9 248 495 804 | 33.70 |
| PHG | Phages | 344 579 387 | 27.37 |
| VRL | Viruses | 3 482 143 321 | 17.09 |
| PLN | Plants | 16 782 598 904 | 14.12 |
| PAT | Patent sequences | 19 219 724 521 | 12.21 |
| SYN | Synthetic | 1 173 218 483 | 12.21 |
| ENV | Environmental samples | 5 590 106 999 | 7.12 |
| MAM | Other mammals | 3 872 932 998 | 6.18 |
| INV | Invertebrates | 17 226 520 457 | 6.07 |
| PRI | Primates | 8 024 647 559 | 2.85 |
| HTC | High-throughput cDNA | 696 583 486 | 2.08 |
| UNA | Unannotated | 208 576 | 1.75 |
| GSS | Genome survey sequences | 25 974 685 352 | 1.08 |
| ROD | Rodents | 4 520 933 672 | 0.42 |
| EST | Expressed sequence tags | 42 640 092 444 | 0.29 |
| HTG | High-throughput genomic | 27 646 512 131 | 0.06 |
| STS | Sequence tagged sites | 640 875 196 | 0.01 |
| TOTAL | All GenBank sequences | 2 635 527 587 818 | 35.52 |
* Measured relative to Release 215 (August 2016)
Figure 1.
Size in base pairs of the five GenBank divisions with the highest annual growth rates in 2017. The growth of GenBank as a whole is also shown as ‘TOTAL’.
Sequence-based taxonomy
Database sequences are classified and can be queried using a comprehensive sequence-based taxonomy (www.ncbi.nlm.nih.gov/taxonomy/) developed by NCBI in collaboration with ENA and DDBJ and with the valuable assistance of external advisers and curators (7,8). About 400 000 formally described species are represented in GenBank, and the top species (not including those in the WGS and TSA divisions) are listed in Table 2.
Table 2. Top Organisms in GenBank.
| Organism | Base pairs* | WGS Genomes** | Non-WGS Genomes** |
|---|---|---|---|
| Homo sapiens | 19 065 856 381 | 58 | 3 |
| Mus musculus | 10 233 714 809 | 21 | 1 |
| Rattus norvegicus | 6 529 312 672 | 9 | 0 |
| Bos taurus | 5 429 768 145 | 2 | 0 |
| Zea mays | 5 228 306 576 | 7 | 0 |
| Sus scrofa | 5 072 476 333 | 15 | 0 |
| Hordeum vulgare | 3 235 943 623 | 7 | 0 |
| Danio rerio | 3 191 032 985 | 3 | 1 |
| Oryzias latipes | 2 836 475 665 | 2 | 3 |
| Ovis canadensis | 2 590 574 434 | 0 | 1 |
| Triticum aestivum | 1 944 658 425 | 12 | 1 |
| Cyprinus carpio | 1 836 551 064 | 1 | 1 |
| Escherichia coli | 1803 951 183 | 8768 | 457 |
| Solanum lycopersicum | 1 746 806 294 | 3 | 1 |
| Oryza sativa | 1 642 593 575 | 18 | 4 |
| Apteryx australis | 1 595 510 956 | 0 | 1 |
| Strongylocentrotus purpuratus | 1 436165 842 | 1 | 0 |
| Macaca mulatta | 1 337 270 420 | 5 | 0 |
| Spirometra erinaceieuropaei | 1 264 448 364 | 0 | 1 |
| Xenopus tropicalis | 1250 011 608 | 1 | 0 |
*Counts correspond to Release 221 and exclude sequences from chloroplasts, mitochondria, metagenomes, uncultured organisms, WGS, and TSA.
**Counts are as of 16 October 2017 and include all INSDC genomes.
Sequence identifiers
Each GenBank record, consisting of both a sequence and its annotations, is assigned a unique identifier called an accession number that is shared across the three collaborating databases (GenBank, DDBJ, ENA). The accession number appears on the ACCESSION line of a GenBank record and remains constant over the lifetime of the record, even when there is a change to the sequence or annotation. Changes to the sequence data itself are tracked by an integer suffix of the accession number, and this Accession.version identifier appears on the VERSION line of the GenBank flat file. Beginning with an initial version of ‘.1’, each change to the sequence data causes the version suffix to increment. The accession portion of the identifier remains unchanged and will always retrieve the most recent version of the record; the older versions remain available under the old accession.version identifiers. The Revision History report, available from the ‘Display Settings’ menu on the default record view in the Nucleotide database (www.ncbi.nlm.nih.gov/nuccore/), summarizes the various updates for a given record, including non-sequence changes. A similar system tracks changes in the corresponding protein translations in the Protein database (www.ncbi.nlm.nih.gov/protein/). These identifiers appear as qualifiers for CDS features in the FEATURES portion of a GenBank entry, e.g. /protein_id = ‘AAF14809.1’.
GenBank uses a somewhat different system of accession.version identifiers for WGS, TSA, and Targeted Loci Study (TLS) sequences. These data are generally submitted as large project sets, and each project is given a ‘master’ record with an accession.version consisting of a four-letter prefix followed by eight zeroes (or nine if the set contains more than one million sequences) and a version suffix. Master records contain no sequence data; rather, they include links to displays of the individual sequences in the Sequence Set Browser (see below). The individual sequence records within a project have accessions consisting of the same four-letter prefix as their master accession, followed by a two-digit version number and a six-digit (or seven-digit) integer ID. For example, the WGS accession number ‘AAAA02002744’ is assigned to sequence number ‘002744’ of the second version of project ‘AAAA’, whose accession number is ‘AAAA00000000.2’. TSA projects have accessions beginning with ‘G’, ‘H’ and ‘I’, while TLS projects have accessions beginning with ‘K’.
Unverified sequences
As reported previously (9), as part of the standard review process for new submissions, GenBank staff may label sequences as unverified if the accuracy of the submitted sequence data or annotations cannot be confirmed. Until the submitter is able to resolve these problems, the definition line of the sequence will begin with ‘UNVERIFIED:’ and the sequence will not be included in BLAST databases. This treatment is being extended to genomic submissions where the source organism is uncertain, there is evidence of contamination, or there are other problems with the data. In addition to the UNVERIFIED label in the definition line, a short description of the problems will be entered in the COMMENT field of the record.
Citing GenBank records
Besides being the primary identifier of a GenBank sequence record, GenBank accession.version identifiers are also the most efficient and reliable way to cite a sequence record in publications. Because searching with a GenBank accession number (without the version suffix) will retrieve the most recent version of a record, the data returned from such searches will change over time if the record is updated. Therefore, sequence data retrieved today by an accession may be different from that discussed or analyzed in a paper published several years ago. We therefore encourage submitters and other authors to include the version suffix when citing a GenBank accession (e.g. AF000001.5), since this ensures that the citation refers to a specific version in time.
BUILDING THE DATABASE
The data in GenBank and the collaborating databases, ENA and DDBJ, are submitted by investigators to one of the three databases. Data are exchanged daily between GenBank, DDBJ and ENA so that daily updates from NCBI servers incorporate the most recently available sequence data from all sources.
Direct electronic submission
Virtually all records enter GenBank as direct electronic submissions (www.ncbi.nlm.nih.gov/genbank/), with the majority of authors using BankIt or the NCBI Submission Portal (submit.ncbi.nlm.nih.gov). Many journals require authors with sequence data to submit the data to a public sequence database as a condition of publication. On average it takes two days for GenBank staff to assign an accession number to a sequence submission, but this can vary depending on the complexity of the submission, with full genomes often requiring more time. GenBank staff assign approximately 3500 accessions per day. The accession number serves as confirmation that the sequence has been submitted and provides a means for readers of articles in which the sequence is cited to retrieve the data. Direct submissions receive a quality assurance review that includes checks for vector contamination, proper translation of coding regions, correct taxonomy, and correct bibliographic citations. A draft of the GenBank record is passed back to the author for review before it enters the database.
Authors may ask that their sequences be kept confidential until the time of publication. Since GenBank policy requires that the deposited sequence data be made public when the sequence or accession number is published, authors are instructed to inform GenBank staff of the publication date of the article in which the sequence is cited in order to ensure a timely release of the data. Although only the submitter is permitted to modify sequence data or annotations, all users are encouraged to report lags in releasing data or possible errors or omissions to GenBank at update@ncbi.nlm.nih.gov.
Submission using BankIt
About a third of author submissions are received through an NCBI web-based data submission tool named BankIt (www.ncbi.nlm.nih.gov/WebSub/?tool=genbank). Using BankIt, authors enter sequence information and biological annotations directly into a series of tabbed forms that allow the submitter to describe the sequence further without having to learn formatting rules or controlled vocabularies. Using BankIt, submitters can submit sets of sequences as well as single sequences. Additionally, BankIt allows submitters to upload source and annotation data using tab-delimited tables. Before creating a draft record in the GenBank flat file format for the submitter to review, BankIt validates the submissions by flagging many common errors and checking for vector contamination using a variant of BLAST called Vecscreen.
Submission using the Submission Portal
The NCBI Submission Portal (submit.ncbi.nlm.nih.gov) is a centralized system that supports submissions of prokaryotic and eukaryotic genomes and a growing number of specialized sequence types, such as ribosomal RNA, TSA, and Sequence Read Archive (SRA). For example, the portal accepts WGS and TSA data in FASTA format using a set of online forms. In addition, the Submission Portal allows submitters to manage BioProject and BioSample submissions while also submitting genome or SRA data. The portal provides links to several submission wizards, help documentation and submission templates. As mentioned above, NCBI continues to add wizards to this interface to assist common submission cases.
Submission using tbl2asn
NCBI works closely with sequencing centers to ensure timely incorporation of bulk data into GenBank for public release. For such large-scale sequencing groups, GenBank offers special batch procedures to facilitate data submission, including the command line program tbl2asn, described at www.ncbi.nlm.nih.gov/genbank/tbl2asn2.html. Using tbl2asn, submitters can convert a table of annotations generated from an annotation pipeline into an ASN.1 record suitable for submission to GenBank. These files for WGS genome and TSA submissions are then transmitted to GenBank through the Submission Portal. A version of tbl2asn called table2asn_GFF also accepts data in the GFF3 format (ftp.ncbi.nih.gov/toolbox/ncbi_tools/converters/by_program/table2asn_GFF).
Notes on particular sequence types
Environmental sample sequences (ENV)
The ENV division of GenBank accommodates sequences obtained using environmental sampling methods in which the sequence is derived directly from the isolate. Records in the ENV division contain ‘ENV’ keywords and use an ‘/environmental_sample’ qualifier in the source feature. Environmental sample sequences are generally submitted for whole metagenomic shotgun sequencing experiments or surveys of sequences from targeted genes, like 16S rRNA. NCBI continues to support BLAST searches (see below) of metagenomic ENV sequences, but sequences within WGS projects are now part of the WGS BLAST database.
Whole genome shotgun sequences
Users should be aware that annotations on WGS project sequences may not be tracked from one assembly version to the next, and so should be considered preliminary. Submitters of genomic sequences, including WGS sequences, are urged to use evidence tags of the form ‘/experimental = text’ and ‘/inference = TYPE:text’, where TYPE is a standard inference type and text consists of structured text. Annotations are not required for complete genomes, but we encourage submitters to request that the genome be annotated by NCBI’s Prokaryotic Genome Annotation Pipeline (www.ncbi.nlm.nih.gov/genome/annotation_prok/) before being released. As part of the bacterial genome submission process, GenBank performs an average nucleotide identity (ANI) analysis to investigate whether the asserted organism name may be incorrect. The analysis compares the submitted genome to all genome assemblies in GenBank from type strains for the reported species. If a new genome has an extremely high ANI and coverage to a type strain from a species other than that reported, GenBank will notify the submitter and press to change the organism name for the submitted genome. Since the analysis uses genomes already in GenBank, it cannot necessarily be performed if GenBank does not have a genome assembly from a type strain for the submitted species.
Transcriptome shotgun assembly (TSA) sequences
The TSA division contains TSA sequences that are assembled from raw sequence reads deposited in the SRA. While SRA is not part of GenBank, it is part of the INSDC and provides access to the data underlying these assemblies (10). TSA records have ‘TSA’ as their keyword and can be retrieved with the query ‘tsa[properties]’ in the Nucleotide database.
Targeted locus studies (TLS)
Targeted locus studies often contain large sets of 16S rRNA sequence or ultra-conserved elements (UCEs). Similar to TSA records, TLS sequences are given a ‘TLS’ keyword and can be retrieved with the query ‘tls[properties]’ in the Nucleotide database. TLS records belong to the appropriate taxonomic GenBank division, and currently all TLS records are in either the VRT, INV or ENV divisions.
Anti-microbial resistance data
As part of the NCBI Pathogen Detection project, NCBI accepts submissions of beta-lactamase sequences as supplementary data for either genome submissions or submissions of novel beta-lactamase sequences (www.ncbi.nlm.nih.gov/pathogens/submit_beta_lactamase/). Beta-lactamase antibiograms should also be submitted, and these will be linked to the BioSample record associated with the submission (www.ncbi.nlm.nih.gov/biosample/docs/beta-lactamase/).
RETRIEVING GENBANK DATA
The Entrez system
The sequence records in GenBank are accessible through the NCBI Entrez retrieval system (11). Records from the EST and GSS divisions of GenBank are stored in the EST and GSS databases, while all other GenBank records are stored in the Nucleotide database. GenBank sequences that are part of population or phylogenetic studies are also collected together in the PopSet database, and conceptual translations of CDS sequences annotated on GenBank records are available in the Protein database. Each of these databases is linked to the scientific literature in PubMed and PubMed Central. Additional information about conducting Entrez searches is found in the NCBI Help Manual (www.ncbi.nlm.nih.gov/books/NBK3831/) and links to related tutorials are provided on the NCBI Learn page (www.ncbi.nlm.nih.gov/home/learn.shtml).
Sequence set browser
As discussed above, a growing number of GenBank records do not have a GI identifier. In such cases, these records are not indexed in Entrez Nucleotide and so cannot be retrieved from the Nucleotide database. For such records, which include many WGS, TLS, and TSA projects, NCBI provides the Sequence Set Browser to support retrieval of these records (www.ncbi.nlm.nih.gov/Traces/wgs/). This interface serves both as a browser that can restrict a list of projects by facets such as taxonomy, source, and BioProject ID, and also as a downloading tool that can provide either metadata tables or actual sequence data from selected projects. While these ‘GI-less’ sequences are not in Entrez Nucleotide, the master records for WGS, TLS, and TSA projects are indexed in Nucleotide and have, at the bottom of their record pages, links to the corresponding set of contigs in the Sequence Set Browser. Protein records derived from these GI-less sequences are included in the new Identical Protein Groups resource (see above), and thus are also accessible through the Entrez system.
Importance of associating sequence records with sequencing projects
NCBI strongly encourages submitters to register large-scale sequencing projects in the BioProject database (www.ncbi.nlm.nih.gov/bioproject). Doing so allows the sequence collection to be represented by a unique project identifier, enabling reliable linkage between sequencing projects and the data they produce. Another benefit is that submitters can include a relevant grant in their BioProject that can then appear in their My Bibliography. A ‘DBLINK’ line appearing in GenBank flat files identifies the sequencing projects associated with a GenBank sequence record. In addition, sequence records may have a link to the BioSample database (12) that provides additional information about the biological materials used in the study. Such studies include genome wide association studies, high-throughput sequencing, microarrays, and epigenomic analyses. As an example, the TSA project GBJS contains DBLINK lines that associate the GenBank sequence record with BioProject record PRJNA255770 and BioSample record SAMN02928618 as well as the two SRA records containing the raw data, SRR1522120 and SRR1522122:
BioProject: PRJNA255770
BioSample: SAMN02928618
Sequence Read Archive: SRR1522120, SRR1522122
While these BioProject identifiers are valuable in representing sequence collections, we would nevertheless recommend that when citing sequence data, as discussed above, it is preferable to use accession.version identifiers to maximize clarity.
In addition to the DBLINK lines for BioProject and BioSample, GenBank records that represent genome assemblies will also have a link to the corresponding record in the Assembly database (13). Assembly records not only collect metadata and statistics for these genome assemblies, but also provide a stable accession for the assembly along with a link to the FTP directory containing the sequence data for the assembly in GenBank, FASTA and GFF3 formats.
BLAST sequence-similarity searching
Sequence-similarity searches are the most fundamental and frequent type of analysis performed on GenBank data. NCBI offers the BLAST family of programs (blast.ncbi.nlm.nih.gov) to detect similarities between a query sequence and database sequences (14,15). BLAST searches may be performed on the NCBI Web site (16) or by using a set of standalone programs distributed by FTP (5). Users should be aware that, because of the enormous diversity of available nucleotide sequence, it is not possible to search all NCBI sequence data at once. Rather, there are several BLAST databases, each suited to a particular type of sequence (Table 3).
Table 3. Selected BLAST nucleotide databases*.
| Database | Contents |
|---|---|
| nt | Taxonomic GenBank divisions |
| env_nt | ENV division |
| tsa_nt | TSA division |
| wgs | WGS sequences |
| 16SMicrobial | Bacterial and archaeal 16S rRNA |
*For more databases, see ftp.ncbi.nlm.nih.gov/blast/documents/blastdb.html
Obtaining GenBank by FTP
NCBI distributes GenBank releases in the traditional flat file format as well as in the ASN.1 format used for internal maintenance. The full bimonthly GenBank release along with daily updates, which incorporate sequence data from ENA and DDBJ, is available by anonymous FTP from NCBI at ftp.ncbi.nlm.nih.gov/genbank. The full release in flat file format is available as a set of compressed files with a non-cumulative set of updates at ftp.ncbi.nlm.nih.gov/genbank/daily-nc/. For convenience in file transfer, the data are partitioned into multiple files; for release 221 there are 2932 files requiring 841 GB of uncompressed disk storage. A script is provided in ftp.ncbi.nlm.nih.gov/genbank/tools/ to convert a set of daily updates into a cumulative update.
MAILING ADDRESS
GenBank, National Center for Biotechnology Information, Building 45, Room 6AN12D-37, 45 Center Drive, Bethesda, MD 20892, USA.
ELECTRONIC ADDRESSES
www.ncbi.nlm.nih.gov - NCBI Home Page.
gb-sub@ncbi.nlm.nih.gov - Submission of sequence data to GenBank.
update@ncbi.nlm.nih.gov - Revisions to, or notification of release of, ‘confidential’ GenBank entries.
info@ncbi.nlm.nih.gov - General information about NCBI resources.
CITING GENBANK
If you use the GenBank database in your published research, we ask that this article be cited.
FUNDING
Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2017; 45:D37–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Toribio A.L., Alako B., Amid C., Cerdeno-Tarraga A., Clarke L., Cleland I., Fairley S., Gibson R., Goodgame N., Ten Hoopen P. et al. European Nucleotide Archive in 2016. Nucleic Acids Res. 2017; 45:D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mashima J., Kodama Y., Fujisawa T., Katayama T., Okuda Y., Kaminuma E., Ogasawara O., Okubo K., Nakamura Y., Takagi T.. DNA Data Bank of Japan. Nucleic Acids Res. 2017; 45:D25–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cochrane G., Karsch-Mizrachi I., Takagi T. International Nucleotide Sequence Database, C. . The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2016; 44:D48–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. NCBI Resource Coordinators Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017; 45:D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014; 42:D7–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012; 40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Federhen S. Type material in the NCBI Taxonomy Database. Nucleic Acids Res. 2015; 43:D1086–D1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benson D.A., Karsch-Mizrachi I., Clark K., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2012; 40:D48–D53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kodama Y., Shumway M., Leinonen R.. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schuler G.D., Epstein J.A., Ohkawa H., Kans J.A.. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996; 266:141–162. [DOI] [PubMed] [Google Scholar]
- 12. Barrett T., Clark K., Gevorgyan R., Gorelenkov V., Gribov E., Karsch-Mizrachi I., Kimelman M., Pruitt K.D., Resenchuk S., Tatusova T. et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012; 40:D57–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kitts P.A., Church D.M., Thibaud-Nissen F., Choi J., Hem V., Sapojnikov V., Smith R.G., Tatusova T., Xiang C., Zherikov A. et al. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 2016; 44:D73–D80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z., Schaffer A.A., Miller W., Madden T.L., Lipman D.J., Koonin E.V., Altschul S.F.. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998; 26:3986–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y. et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013; 41:W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]