Abstract
PolyA_DB is a database cataloging cleavage and polyadenylation sites (PASs) in several genomes. Previous versions were based mainly on expressed sequence tags (ESTs), which had a limited amount and could lead to inaccurate PAS identification due to the presence of internal A-rich sequences in transcripts. Here, we present an updated version of the database based solely on deep sequencing data. First, PASs are mapped by the 3′ region extraction and deep sequencing (3′READS) method, ensuring unequivocal PAS identification. Second, a large volume of data based on diverse biological samples increases PAS coverage by 3.5-fold over the EST-based version and provides PAS usage information. Third, strand-specific RNA-seq data are used to extend annotated 3′ ends of genes to obtain more thorough annotations of alternative polyadenylation (APA) sites. Fourth, conservation information of PAS across mammals sheds light on significance of APA sites. The database (URL: http://www.polya-db.org/v3) currently holds PASs in human, mouse, rat and chicken, and has links to the UCSC genome browser for further visualization and for integration with other genomic data.
INTRODUCTION
Cleavage and polyadenylation (C/P) of the nascent RNA is essential for 3′ end maturation of almost all eukaryotic mRNAs and long non-coding RNAs (ncRNAs), and preludes termination of transcription (1,2). The C/P site, also known as polyA site (PAS), is defined by multiple surrounding regulatory cis elements (3). In vertebrates, the cis elements include the PAS hexamer (AAUAAA, AUUAAA, or their variants), UGUA motif and U-rich motifs, all located upstream of the PAS, and downstream U-rich and UGUG motifs. PAS cis elements vary in lower species (3–6), with the budding yeast showing the most degenerate motifs around the PAS (3,7).
Most eukaryotic genes harbor multiple PASs, leading to expression of alternative polyadenylation (APA) isoforms (1,8,9). Most APA sites are located in 3′ untranslated regions (3′UTRs) of mRNAs, resulting in isoforms with different 3′UTR lengths and, consequently, distinct mRNA metabolisms. In addition, a sizable fraction of the sites are embedded in introns (10,11), influencing both coding and non-coding regions of gene transcripts. APA greatly increases the diversity of transcriptome encoded by a genome, and has been shown to be highly regulated across tissues and cell types (12–14). In addition, global regulation of the APA profile has been shown in cell proliferation, differentiation, and development (15,16), and in cells responding to environmental cues (17,18).
Given the critical role of PAS in termination of transcription and the impact of APA on gene expression, it is important to have a comprehensive and accurate catalog of PASs in genomes. Early PAS databases, such as PolyA_DB (19,20) and PACdb (21), were based on cDNA sequences and expressed sequence tags (ESTs). PASs were identified using cDNA/EST sequences that had an terminal poly(A/T) region corresponding to the poly(A) tail (22,23). While these databases were useful for initial understanding of the scale of APA and facilitated survey-based analysis of APA in different systems, they are not comprehensive due to the limited number of cDNA/EST sequences available in public databases. In addition, internal A-rich sequences of transcripts often lead to poly(A/T) sequences in cDNAs, resulting in false identification of PAS (24).
The last decade has witnessed explosive growth of deep sequencing (a.k.a., next-generation sequencing) data. A number of sequencing methods have been developed to specifically interrogate the 3′ end of transcripts (reviewed in (25)), which have also led to the creation of several PAS-based databases, such as APADB (26) and APASdb (27). However, while 3′ end sequencing methods have greatly facilitated PAS identification genome-wide, priming at internal A-rich sequences is still an issue leading to false identification of PASs when an oligo(dT)-containing primer is used to generate cDNAs (22). Whereas false positives come from internal A-rich sequences, false negatives arise when genuine PASs are discarded because of their placement in an A-rich sequence region (28).
Here we present a major upgrade of PolyA_DB (named version 3), built upon a large volume of data generated by 3′READS (11,28), a 3′ end sequencing method that is not affected by internal A-rich sequences. At the time of this publication, the database contains PASs in four organisms, human, mouse, rat and chicken. Conservation of PAS across mammals and transcript abundance for each PAS provide additional information to examine the relative importance of APA sites.
MATERIALS AND METHODS
Identification of PASs with 3′READS data
3′READS is a deep sequencing method specialized in interrogation of the 3′ end of poly(A)+ transcripts (11,28). The method uses a chimeric oligo containing DNA and RNA (or locked nucleic acid) to retain the 5′ end region of poly(A) tail in the cDNA (11,28). As such, each 3′READS read contains a few terminal T’s (because of the antisense sequencing of cDNA) corresponding to the poly(A) tail. We collected over 9–59 3′READS samples per species from diverse tissues and cell lines of human, mouse, rat and chicken, totaling 23–150 million PAS-containing reads per species (Table 1 and Supplementary Table S1). We aligned reads to corresponding genomes (mm9 for mouse, hg19 for human, rn5 for rat and galGal4 for chicken) for identification of PASs using bowtie2 (version 2.2.9). Random nucleotides at the 5′ end (derived from the 3′ adapter used for cDNA construction) of reads were removed before mapping. Reads with a mapping quality score (MAPQ) ≥10 were kept for further analysis. Reads with ≥2 non-genomic 5′Ts after alignment were called PAS reads (11). As such, internal A-rich sequences of transcripts, which would result in reads without extra T’s after genome alignment, did not affect PAS identification. For each sample, the PASs within 24 nt from each other were clustered to address heterogeneous cleavage in PAS usage (29). Only the PASs with at least two reads in at least two samples were considered as genuine PASs.
Table 1. Summary of PolyA_DB version 3.1.
| Species | Human | Mouse | Rat | Chicken |
|---|---|---|---|---|
| No. of samples used | 24 | 59 | 11 | 9 |
| No. of PAS reads used | 59 090 907 | 153 989 213 | 23 616 600 | 29 104 491 |
| No. of PASs | 108 042 | 202 426 | 61 905 | 65 909 |
| No. of genic PASs | 85 275 | 121 163 | 36 941 | 45 116 |
| No. of genes listed | 20 998 | 21 588 | 14 529 | 12 292 |
| No. of genes with 3′ end extension | 8962 | 12 027 | 8302 | 8352 |
| Median 3′ end extension size (nt) | 758 | 469 | 617 | 1062 |
| No. of mRNA genes | 15 977 | 17 846 | 14 077 | 12 130 |
| No. of ncRNA genes | 5021 | 3742 | 452 | 162 |
PAS annotation
Identified PASs were assigned to genes based on RefSeq database (Release 83) (30) and Ensembl database (release 75 for human, release 67 for mouse, release 79 for rat and release 85 for chicken) (31). Because RefSeq and Ensembl gene annotations often miss PASs at the 3′ end of genes, we used strand-specific, poly(A)+ RNA-seq datasets (32–36) to extend the 3′ ends defined by RefSeq and Ensembl. We required continuous coverage of RNA-seq reads in the extended region, with a minimum of five reads at each position. We also required that 3′ end extension did not exceed the transcription start site of the downstream gene on the same strand. We then annotated genic PASs by their intron/exon locations based on the representative RefSeq or Ensembl sequences (the sequence with greatest genomic span), i.e. 5′-most exon, internal exon, 3′-most exon, single exon and intron. This step was carried out for both mRNA and ncRNA genes. When a gene was annotated in both RefSeq and Ensembl databases, RefSeq information was used.
For mRNA genes, we next classified PASs into four types based on coding information derived from the representative RefSeq or Ensembl sequence, including 5′UTR, CDS, 3′UTR and intron. Because most 3′UTRs harbor multiple PASs, we further classified 3′UTR PASs into first, middle and last PASs, based their relative locations. If a gene had a single 3′UTR PAS, it was called single PAS. Moreover, we annotated the PAS hexamer sequence for each PAS, using the 40-nt upstream region of the PAS (29). Five types were included, i.e. AAUAAA, AUUAAA, Other (AGUAAA UAUAAA CAUAAA GAUAAA AAUAUA AAUACA AAUAGA AAAAAG ACUAAA), A-rich (AAAAAA) and None.
Conservation of PASs
We used pair-wise genome alignment chain files from the UCSC Genome Bioinformatics Site to obtain syntenic regions between genomes. We used the reciprocal best match method our lab previously developed to identify conserved PASs (37). Briefly, two PASs from two species were considered to be orthologous when they were within 24-nt from one another in whole genome alignment. A PAS that is conserved between any two of the three analyzed mammals (human, mouse and rat) was considered as a conserved site in the current release.
PAS usage levels
To evaluate the usage level of each PAS, we developed two metrics, percentage of samples expressed (PSE) and mean RPM (reads per million), based on the samples we used for 3′READS. The PSE of a PAS was calculated as NExpressed/NTotal, where NExpressed is the number of samples in which the usage of PAS was detected (≥2 reads per sample), and NTotal is the total number of samples used. The mean RPM of each PAS is averaged RPM value across all the samples in which its usage was detected (≥2 reads). The RPM value of a PAS in each sample is the number of reads for the PAS normalized to the total number of reads mapped to the genome.
DATABASE CONTENT
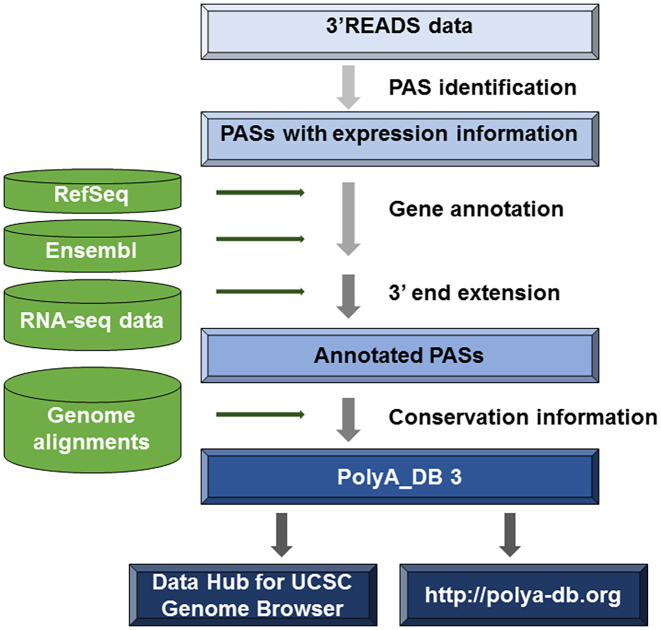
The approach for PAS identification and presentation in PolyA_DB version 3 is summarized in Figure 1. As of September 2017, PolyA_DB version 3 contains 85 275, 121 163, 36 941 and 45 116 genic PASs covering 20 998 human, 21 588 mouse, 14 529 rat and 12 292 chicken genes, respectively (Table 1). The PAS coverage is significantly higher than the previous version, PolyA_DB 2, with an overall increase by 3.5-fold (1.6-fold for human, 4.0-fold for mouse, 1.4-fold for rat and 7.2-fold for chicken).
Figure 1.
Schematic of PAS identification and presentation in PolyA_DB version 3. The data flow is indicated by arrowed lines. See the main text for details.
PolyA_DB substantially improves 3′ end annotations of genes in RefSeq and Ensembl databases. The 3′ end of each gene was extended by public strand-specific RNA-seq data and the PASs identified by 3′READS (see Materials and Methods). Overall, ∼56% of genes (both mRNA and ncRNA genes) had 3′ end extension. The median extension size ranged from 469- to 1062-nt (Table 1).
PASs in PolyA_DB are annotated according the splicing configuration derived from representative sequences in RefSeq and Ensembl databases (see ‘Materials and Methods’ section). This process was carried out for both mRNA and ncRNA genes. For mRNA genes (Supplementary Table S2), we further classified PASs according to the coding region. In all the species analyzed, about 66–81% of the genic PASs were located in 3′UTRs, followed by intronic PASs (17–32%), which would change both CDS and 3′UTR. For ncRNA genes in mammals (Supplementary Table S3), more than half of their PASs were found in 3′-most exons (including single exon genes), followed by introns (21–42%) and internal exons (3–7%).
Each PAS in PolyA_DB is annotated with two types of information that reflects its usage levels, including frequency of detection of its usage across samples and average expression level (number of normalized reads) in the samples it was detected. In addition, PAS hexamer sequence is shown to indicate PAS strength, and conservation in mammals (human, mouse and rat) is displayed to help understand the evolutionary importance of PAS.
DATA ACCESS AND WEBSITE INTERFACE
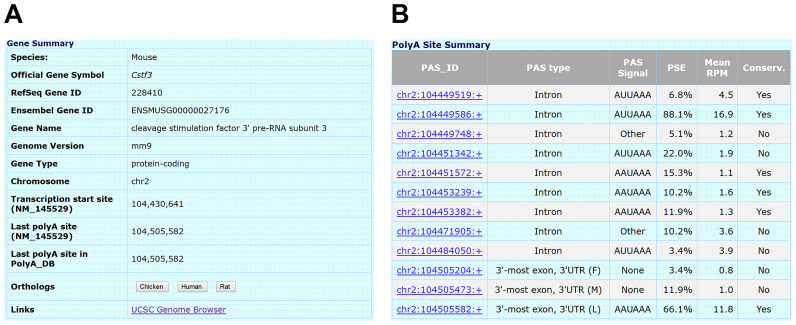
Data in PolyA_DB are stored in a relational database, implemented with MySQL (38). The interactive web interface is implemented with PHP (URL: http://www.polya-db.org/v3). Queries are based on RefSeq gene symbol/ID or Ensembl gene ID. We provide two view tables to show data: the Gene view table (Figure 2A) provides a summary of the queried gene, including gene symbol, gene ID (both RefSeq and Ensembl), gene name, gene type, genome version and annotated transcription start site and the last PASs based on RefSeq or Ensembl and PolyA_DB. Orthologous genes in other species in PolyA_DB are listed, which are based on the HomoloGene database from NCBI. Finally, a link to UCSC genome browser is provided for visualization of the gene and for integration with other public genomic data.
Figure 2.
An example of search result from PolyA_DB 3. (A) Gene view. Mouse gene Cstf3 is used as an example. The output includes a summary table of the gene as well as a link to UCSC genome browser. (B) PolyA SiteView. This table contains information of all individual PASs assigned to the queried gene and their links to UCSC genome browser.
The PolyA Site View table (Figure 2B) lists all the PASs assigned to the queried gene. For each PAS, we provide information about its genomic location (also used as ID for the PAS, or PAS_ID), intron/exon location (5′-most exon, 3′-most exon, internal exon and intron), PAS type (5′UTR, CDS, 3′UTR and Intron), PSE, mean RPM and conservation in mammals. A link to UCSC genome browser is also provided for each PAS.
The PolyA_DB data can also be viewed on UCSC genome browser through a custom track. The URL for the PolyA_DB track hub is http://www.polya-db.org/v3/hub/. As in PolyA_DB, each PAS is identified by its PAS_ID with conservation information (‘C’ for conserved, ‘N’ for non-conserved). The mean RPM of all samples can also be displayed. In addition, batch download of data in a tabular format is available at http://www.polya-db.org/v3/download.
SUMMARY AND FUTURE DIRECTIONS
Here, we present a major upgrade of PolyA_DB (version 3), which substantially expands PAS collections in several species. With accurate PAS identification and quantitative usage data based on a large number of samples, as well as conservation information across species, PolyA_DB 3 will be of use for 3′ end annotation of genes and for understanding the significance of APA sites. Future work will add data from more species and more diverse cell/tissue types, which will help APA conservation and regulation studies. With more RNA-seq and 3′READS data becoming available, we also expect a more precise definition of the 3′ end of genes in the future. In addition, in-depth analysis of PASs in introns and in intergenic regions will be carried out to elucidate their functions in gene regulation and contributions to transcriptional activities in the genome.
Supplementary Material
ACKNOWLEDGEMENTS
We thank other members of our laboratory for helpful discussions and database testing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [GM084089 to B.T.]. Funding for open access charge: NIH [GM084089].
Conflict of interest statement. None declared.
REFERENCES
- 1. Tian B., Manley J.L.. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017; 18:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Proudfoot N.J. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science. 2016; 352:aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tian B., Graber J.H.. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip. Rev. RNA. 2012; 3:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jan C.H., Friedman R.C., Ruby J.G., Bartel D.P.. Formation, regulation and evolution of Caenorhabditis elegans 3΄UTRs. Nature. 2011; 469:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haenni S., Ji Z., Hoque M., Rust N., Sharpe H., Eberhard R., Browne C., Hengartner M.O., Mellor J., Tian B. et al. Analysis of C. elegans intestinal gene expression and polyadenylation by fluorescence-activated nuclei sorting and 3΄-end-seq. Nucleic Acids Res. 2012; 40:6304–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Graber J.H., Cantor C.R., Mohr S.C., Smith T.F.. In silico detection of control signals: mRNA 3΄-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:14055–14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu X., Hoque M., Larochelle M., Lemay J.F., Yurko N., Manley J.L., Bachand F., Tian B.. Comparative analysis of alternative polyadenylation in S. cerevisiae and S. pombe. Genome Res. 2017; 27:1685–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012; 18:2105–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elkon R., Ugalde A.P., Agami R.. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013; 14:496–506. [DOI] [PubMed] [Google Scholar]
- 10. Tian B., Pan Z., Lee J.Y.. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007; 17:156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoque M., Ji Z., Zheng D., Luo W., Li W., You B., Park J.Y., Yehia G., Tian B.. Analysis of alternative cleavage and polyadenylation by 3΄ region extraction and deep sequencing. Nat. Methods. 2013; 10:133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang H., Lee J.Y., Tian B.. Biased alternative polyadenylation in human tissues. Genome Biol. 2005; 6:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B.. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lianoglou S., Garg V., Yang J.L., Leslie C.S., Mayr C.. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013; 27:2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ji Z., Lee J.Y., Pan Z., Jiang B., Tian B.. Progressive lengthening of 3΄ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B.. Proliferating cells express mRNAs with shortened 3΄ untranslated regions and fewer microRNA target sites. Science. 2008; 320:1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Flavell S.W., Kim T.-K., Gray J.M., Harmin D.A., Hemberg M., Hong E.J., Markenscoff-Papadimitriou E., Bear D.M., Greenberg M.E.. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008; 60:1022–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang J.-W., Zhang W., Yeh H.-S., De Jong E.P., Jun S., Kim K.-H., Bae S.S., Beckman K., Hwang T.H., Kim K.-S.. mRNA 3΄-UTR shortening is a molecular signature of mTORC1 activation. Nat. Commun. 2015; 6:7218. [DOI] [PubMed] [Google Scholar]
- 19. Zhang H., Hu J., Recce M., Tian B.. PolyA_DB: a database for mammalian mRNA polyadenylation. Nucleic Acids Res. 2005; 33:D116–D120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee J.Y., Yeh I., Park J.Y., Tian B.. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 2007; 35:D165–D168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brockman J.M., Singh P., Liu D., Quinlan S., Salisbury J., Graber J.H.. PACdb: polyA cleavage site and 3′-UTR database. Bioinformatics. 2005; 21:3691–3693. [DOI] [PubMed] [Google Scholar]
- 22. Gautheret D., Poirot O., Lopez F., Audic S., Claverie J.-M.. Alternate polyadenylation in human mRNAs: a large-scale analysis by EST clustering. Genome Res. 1998; 8:524–530. [DOI] [PubMed] [Google Scholar]
- 23. Lee J.Y., Park J.Y., Tian B.. Identification of mRNA polyadenylation sites in genomes using cDNA sequences, expressed sequence tags, and Trace. Methods Mol. Biol. 2008; 419:23–37. [DOI] [PubMed] [Google Scholar]
- 24. Nam D.K., Lee S., Zhou G., Cao X., Wang C., Clark T., Chen J., Rowley J.D., Wang S.M.. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:6152–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng D., Tian B.. RNA-binding proteins in regulation of alternative cleavage and polyadenylation. Adv. Exp. Med. Biol. 2014; 825:97–127. [DOI] [PubMed] [Google Scholar]
- 26. Muller S., Rycak L., Afonso-Grunz F., Winter P., Zawada A.M., Damrath E., Scheider J., Schmah J., Koch I., Kahl G. et al. APADB: a database for alternative polyadenylation and microRNA regulation events. Database. 2014; 2014:bau076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. You L., Wu J., Feng Y., Fu Y., Guo Y., Long L., Zhang H., Luan Y., Tian P., Chen L.. APASdb: a database describing alternative poly (A) sites and selection of heterogeneous cleavage sites downstream of poly (A) signals. Nucleic Acids Res. 2014; 43:D59–D67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng D., Liu X., Tian B.. 3΄READS+, a sensitive and accurate method for 3΄ end sequencing of polyadenylated RNA. RNA. 2016; 22:1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tian B., Hu J., Zhang H., Lutz C.S.. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005; 33:201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pruitt K.D., Tatusova T., Maglott D.R.. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2006; 35:D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al. Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuo R.I., Tseng E., Eory L., Paton I.R., Archibald A.L., Burt D.W.. Normalized long read RNA sequencing in chicken reveals transcriptome complexity similar to human. BMC Genomics. 2017; 18:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mason A.S., Fulton J.E., Hocking P.M., Burt D.W.. A new look at the LTR retrotransposon content of the chicken genome. BMC Genomics. 2016; 17:688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Merkin J., Russell C., Chen P., Burge C.B.. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012; 338:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pervouchine D.D., Djebali S., Breschi A., Davis C.A., Barja P.P., Dobin A., Tanzer A., Lagarde J., Zaleski C., See L.H. et al. Enhanced transcriptome maps from multiple mouse tissues reveal evolutionary constraint in gene expression. Nat. Commun. 2015; 6:5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J.Y., Ji Z., Tian B.. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3΄-end of genes. Nucleic Acids Res. 2008; 36:5581–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Welling L., Thomson L.. PHP and MySQL Web Development. 2003; 5th ednBoston, Massachusetts: Addison-Wesley Professional. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.