Abstract
Accumulating evidence suggests that diverse non-coding RNAs (ncRNAs) are involved in the progression of a wide variety of diseases. In recent years, abundant ncRNA–disease associations have been found and predicted according to experiments and prediction algorithms. Diverse ncRNA–disease associations are scattered over many resources and mammals, whereas a global view of diverse ncRNA–disease associations is not available for any mammals. Hence, we have updated the MNDR v2.0 database (www.rna-society.org/mndr/) by integrating experimental and prediction associations from manual literature curation and other resources under one common framework. The new developments in MNDR v2.0 include (i) an over 220-fold increase in ncRNA–disease associations enhancement compared with the previous version (including lncRNA, miRNA, piRNA, snoRNA and more than 1400 diseases); (ii) integrating experimental and prediction evidence from 14 resources and prediction algorithms for each ncRNA–disease association; (iii) mapping disease names to the Disease Ontology and Medical Subject Headings (MeSH); (iv) providing a confidence score for each ncRNA–disease association and (v) an increase of species coverage to six mammals. Finally, MNDR v2.0 intends to provide the scientific community with a resource for efficient browsing and extraction of the associations between diverse ncRNAs and diseases, including >260 000 ncRNA–disease associations.
INTRODUCTION
Mammalian genomes produce many thousands of regulatory ncRNAs that are involved in a variety of biological functions (1–3). The identification of roles of ncRNAs in the genesis and progression of pathological disorders is booming (4–6). Over the past decade, many experimental techniques and prediction algorithms were developed, leading to the expansion of many diverse ncRNA–disease association datasets. Consequently, several databases that document the relevance of lncRNA/miRNA to diseases have been constructed, providing useful results with experimental evidence from the literature (7–9). Some computational algorithms also focus on predicting lncRNA/miRNA–disease associations according to the sequence- or path-based computational model (10–13). In addition, other ncRNAs, such as piRNAs and snoRNAs, have also been demonstrated to contribute to diseases (1,14). Although diverse ncRNA–disease associations are scattered over various resources and mammals, a global view of diverse ncRNA–disease associations is not available for any particular mammal.
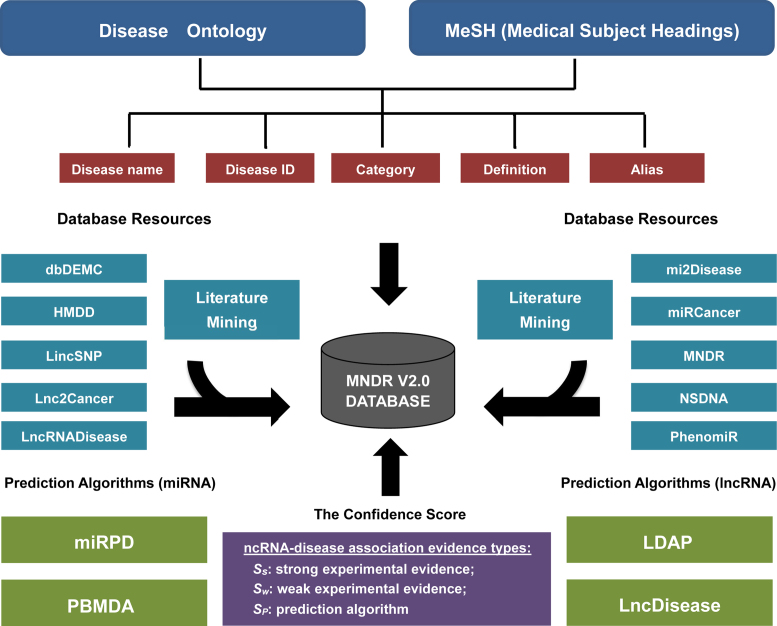
Hence, we updated the MNDR v2.0 database (http://www.rna-society.org/mndr/) by integrating manual literature curation, 14 experimental resources and prediction algorithms (Figure 1). Accordingly, MNDR v2.0 offers several distinct advantages from its first release database: (i) integration from manual literature curation, 10 experimental resources and 4 prediction algorithms, recruiting >260 000 ncRNA–disease associations with >1400 diseases, exceeding a 220-fold increase over the previous version; (ii) providing a confidence score for each ncRNA–disease association; (iii) mapping disease names to the Disease Ontology (15) and MeSH (16) and (iv) an increase of species coverage to six mammals (Homo sapiens, Macaca mulatta, Mus musculus, Pan troglodytes, Rattus norvegicus and Sus scrofa). MNDR v2.0 allows users to query, analyze and manipulate mammalian ncRNA–disease knowledge and provides a valuable resource for investigating the hidden associations between diverse ncRNAs and diseases.
Figure 1.
Overview of the MNDR v2.0 database.
DATA COLLECTION
To update the MNDR v2.0 database, we first screened over 26 600 studies within the PubMed database (mainly from 2012 to 2017) with the following keyword combinations: (ncRNA symbols or ncRNA category names), (mammal names) and (disease names). Three major types of ncRNA symbols were used: (i) lncRNA symbols collected from the lncRNAdb (17) and Ensembl databases (18), (ii) miRNA symbols collected from the miRBase database (19), (iii) snoRNAs symbols collected from the sno/scaRNAbase (20) and snoRNA-LBME-db (21). The lists of disease terms were collected according to the Disease Ontology (15) and MeSH (16) vocabularies. The relevant hits were downloaded and prepared systematically for further manual data curation. Then, we recovered 6033 ncRNA–disease associations in these studies. Second, MNDR v2.0 integrated diverse ncRNA–disease associations from another 10 experimental resources, including dbDEMC (22), HMDD (23), Lnc2Cancer (24), LincSNP (25), LncRNADisease (8), miR2Disease (7), miRCancer (26), MNDR (9), NSDNA (27) and PhenomiR (28) and four prediction algorithms (LDAP (12), LncDisease (13), miRPD (10) and PBMDA (11)).
DATABASE CONTENT AND CONSTRUCTION
In total, MNDR v2.0 contains 8824 experimental lncRNA-associated, 70 381 experimental miRNA-associated, 118 experimental piRNA-associated and 67 experimental snoRNA-associated entries across 6 mammals (70 404 experimental Homo sapiens-associated, 63 experimental Macaca mulatta-associated, 6218 experimental Mus musculus-associated, 45 experimental Pan troglodyte-associated, 2549 experimental R. norvegicus-associated and 111 experimental Sus scrofa-associated entries) and documents 11 504 published studies (Figure 2). According to four prediction algorithms, MNDR v2.0 also includes 153 508 predicted lncRNA-associated, and 28 144 predicted miRNA-associated entries for Homo sapiens (61 592 lncRNA-associated entries predicted by LDAP, 96 369 lncRNA-associated entries predicted by LncDisease, 12 140 miRNA-associated entries predicted by miRPD and 16 738 miRNA-associated entries predicted by PBMDA) (Figure 2). Among these ncRNA–disease associations, it contains 19 575 non-redundant lncRNAs, 4150 non-redundant miRNAs, 110 non-redundant piRNAs and 23 non-redundant snoRNAs associated with 1416 disease terms.
Figure 2.
Statistics of diverse ncRNA–disease associations according to RNA categories and mammals. (A) The percentage of diverse ncRNA–disease associations in the MNDR v2.0 database; (B) The number of ncRNA–disease associations according to mammals in the MNDR v2.0 database, and the height of histogram transformed by log10.
In MNDR v2.0, each ncRNA–disease association contains detailed information, including confidence score, ncRNA symbol, ncRNA ID/miRBase accession, ncRNA category, species, related target gene, disease detail information, evidence support, PubMed ID, and description. To facilitate researchers accessing information from external resources, we linked lncRNA symbols to the Ensembl or NCBI Gene or Nucleotide database, miRNA symbols to the miRBase database (19), and other ncRNAs to the NCBI Gene database. More importantly, considering disease names collected from different resources, MNDR v2.0 presents the standard architecture system for disease according to Disease Ontology (15) and MeSH (16), including standard disease name, disease id, category, definition and alias.
On the updated ‘Search’ page, the confidence score of each ncRNA–disease association is added in the ‘Advanced Filter’ option, where users can select ncRNA–disease associations by a user-specific threshold. On the updated ‘Browse’ page, users can access MNDR v2.0 via three different paths: ‘Diseases’, ‘ncRNA Category’ and ‘Species’. To complete the data of ncRNA–disease associations, MNDR v2.0 allows researchers to submit established diverse ncRNA–disease association entries that are not documented. In addition, all diverse ncRNA–disease associations can be downloaded directly in the TXT format, and MNDR v2.0 provides a publicly available interface (API) for automatic data retrieval in the ‘Download & API’ page.
CONFIDENCE SCORE
In MNDR v2.0, diverse ncRNA–disease associations are collected from different experimental resources and prediction algorithms. Similar to miRTarBase (29) and the RAID v2.0 database (30), experimental evidence in MNDR v2.0 is classified into strong and weak evidence by manual assignment. By integrating the experimental and prediction evidence, we developed a confidence score system to evaluate the reliability of a specific ncRNA–disease association that combines scores from all of these independent evidence resources. In principle, experimental evidence should contribute more importantly to the confidence score than prediction evidence; strong experimental evidence should provide more reliable evidence than weak experimental evidence; and ncRNA–disease associations supported by more evidence should be given significantly higher confidence scores than those supported by weaker evidence. Similar to the RAID v2.0 database (30), according to the evidence types and number of evidence resources, we calculate the confidence score (S) for each ncRNA–disease association as follows:
 |
(1) |
where i is the evidence type (s: strong experimental evidence, w: weak experimental evidence, p: prediction algorithm), x is the number of evidence resources, we set weight factor ws, ww and wp to 1, 0.65 and 0.15, respectively (if x = 0, we set weight factor wi to 0). Only well-supported ncRNA–disease associations obtain a value close to 1 (score ranges between 0 and 1). Therefore, this is an effective tool for filtering reliable ncRNA–disease associations.
CONCLUSION
In the past decade, substantial studies have explored numerous ncRNAs involved in the genesis and progression of pathological disorders. Currently, the comprehensive understanding of the mutual regulations among diverse ncRNAs in diseases remains ambiguous in mammals. Consequently, we have updated the MNDR v2.0 database by integrating manual literature curation and 14 resources/prediction algorithms under one common framework. The aim is to provide a comprehensive and reliably assessed collection of ncRNA–disease associations in mammals. Hence, MNDR v2.0 will be of particular interest to the life science community and facilitates the biologists to unravel the role of diverse ncRNAs in the pathogenesis of diseases. Importantly, new rational drug target design and prognosis biomarker development will benefit from a clear understanding of diverse ncRNAs-mediated disease network.
FUNDING
National Natural Science Foundation of China [81770104]; Natural Science Foundation of Heilongjiang Province [C2015027]; WeihanYu Youth Science Fund Project of Harbin Medical University and Graduate Training Programs for Innovation [YJSCX2015-44HYD]. Funding for open access charge: National Natural Science Foundation of China [81770104]; Natural Science Foundation of Heilongjiang Province [C2015027]; WeihanYu Youth Science Fund Project of Harbin Medical University and Graduate Training Programs for Innovation [YJSCX2015-44HYD].
Conflict of interest statement. None declared.
REFERENCES
- 1. Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011; 12:861–874. [DOI] [PubMed] [Google Scholar]
- 2. Schwarzer A., Emmrich S., Schmidt F., Beck D., Ng M., Reimer C., Adams F.F., Grasedieck S., Witte D., Kabler S. et al. The non-coding RNA landscape of human hematopoiesis and leukemia. Nat. Commun. 2017; 8:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coffre M., Koralov S.B.. miRNAs in B cell development and lymphomagenesis. Trends Mol. Med. 2017; 23:721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cooper T.A., Wan L., Dreyfuss G.. RNA and disease. Cell. 2009; 136:777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Uchida S., Dimmeler S.. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 2015; 116:737–750. [DOI] [PubMed] [Google Scholar]
- 6. Mendell J.T., Olson E.N.. MicroRNAs in stress signaling and human disease. Cell. 2012; 148:1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y.. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009; 37:D98–D104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q.. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013; 41:D983–D986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y., Chen L., Chen B., Li X., Kang J., Fan K., Hu Y., Xu J., Yi L., Yang J. et al. Mammalian ncRNA–disease repository: a global view of ncRNA-mediated disease network. Cell Death Dis. 2013; 4:e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mork S., Pletscher-Frankild S., Palleja Caro A., Gorodkin J., Jensen L.J.. Protein-driven inference of miRNA–disease associations. Bioinformatics. 2014; 30:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. You Z.H., Huang Z.A., Zhu Z., Yan G.Y., Li Z.W., Wen Z., Chen X.. PBMDA: A novel and effective path-based computational model for miRNA–disease association prediction. PLoS Comput. Biol. 2017; 13:e1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lan W., Li M., Zhao K., Liu J., Wu F.X., Pan Y., Wang J.. LDAP: a web server for lncRNA–disease association prediction. Bioinformatics. 2017; 33:458–460. [DOI] [PubMed] [Google Scholar]
- 13. Wang J., Ma R., Ma W., Chen J., Yang J., Xi Y., Cui Q.. LncDisease: a sequence based bioinformatics tool for predicting lncRNA–disease associations. Nucleic Acids Res. 2016; 44:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sana J., Faltejskova P., Svoboda M., Slaby O.. Novel classes of non-coding RNAs and cancer. J. Transl. Med. 2012; 10:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kibbe W.A., Arze C., Felix V., Mitraka E., Bolton E., Fu G., Mungall C.J., Binder J.X., Malone J., Vasant D. et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015; 43:D1071–D1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bhattacharya S., Ha-Thuc V., Srinivasan P.. MeSH: a window into full text for document summarization. Bioinformatics. 2011; 27:i120–i128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quek X.C., Thomson D.W., Maag J.L., Bartonicek N., Signal B., Clark M.B., Gloss B.S., Dinger M.E.. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015; 43:D168–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al. Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xie J., Zhang M., Zhou T., Hua X., Tang L., Wu W.. Sno/scaRNAbase: a curated database for small nucleolar RNAs and cajal body-specific RNAs. Nucleic Acids Res. 2007; 35:D183–D187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lestrade L., Weber M.J.. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006; 34:D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang Z., Wu L., Wang A., Tang W., Zhao Y., Zhao H., Teschendorff A.E.. dbDEMC 2.0: updated database of differentially expressed miRNAs in human cancers. Nucleic Acids Res. 2017; 45:D812–D818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Y., Qiu C., Tu J., Geng B., Yang J., Jiang T., Cui Q.. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014; 42:D1070–D1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ning S., Zhang J., Wang P., Zhi H., Wang J., Liu Y., Gao Y., Guo M., Yue M., Wang L. et al. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016; 44:D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ning S., Yue M., Wang P., Liu Y., Zhi H., Zhang Y., Zhang J., Gao Y., Guo M., Zhou D. et al. LincSNP 2.0: an updated database for linking disease-associated SNPs to human long non-coding RNAs and their TFBSs. Nucleic Acids Res. 2017; 45:D74–D78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie B., Ding Q., Han H., Wu D.. miRCancer: a microRNA-cancer association database constructed by text mining on literature. Bioinformatics. 2013; 29:638–644. [DOI] [PubMed] [Google Scholar]
- 27. Wang J., Cao Y., Zhang H., Wang T., Tian Q., Lu X., Lu X., Kong X., Liu Z., Wang N. et al. NSDNA: a manually curated database of experimentally supported ncRNAs associated with nervous system diseases. Nucleic Acids Res. 2017; 45:D902–D907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ruepp A., Kowarsch A., Theis F.. PhenomiR: microRNAs in human diseases and biological processes. Methods Mol. Biol. 2012; 822:249–260. [DOI] [PubMed] [Google Scholar]
- 29. Chou C.H., Chang N.W., Shrestha S., Hsu S.D., Lin Y.L., Lee W.H., Yang C.D., Hong H.C., Wei T.Y., Tu S.J. et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016; 44:D239–D247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yi Y., Zhao Y., Li C., Zhang L., Huang H., Li Y., Liu L., Hou P., Cui T., Tan P. et al. RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic Acids Res. 2017; 45:D115–D118. [DOI] [PMC free article] [PubMed] [Google Scholar]