Abstract
Enhancers, as specialized genomic cis-regulatory elements, activate transcription of their target genes and play an important role in pathogenesis of many human complex diseases. Despite recent systematic identification of them in the human genome, currently there is an urgent need for comprehensive annotation databases of human enhancers with a focus on their disease connections. In response, we built the Human Enhancer Disease Database (HEDD) to facilitate studies of enhancers and their potential roles in human complex diseases. HEDD currently provides comprehensive genomic information for ∼2.8 million human enhancers identified by ENCODE, FANTOM5 and RoadMap with disease association scores based on enhancer–gene and gene–disease connections. It also provides Web-based analytical tools to visualize enhancer networks and score enhancers given a set of selected genes in a specific gene network. HEDD is freely accessible at http://zdzlab.einstein.yu.edu/1/hedd.php.
INTRODUCTION
Enhancers are specialized genomic cis-regulatory elements, capable of activating transcription of their target genes at great distances, and play a central role in regulating a wide range of important biological functions and processes, such as embryogenesis, development, and homeostasis, whose impairment could result in diseases (1). Numerous studies have shown that genetic variants associated with human complex diseases are significantly enriched in transcription-factor-occupied regions and DNase I hypersensitive sites, most of which overlap with enhancer regions (2–5). For example, SNPs associated with Type 2 diabetes are highly enriched in the pancreatic islet clustered enhancers, and ∼88% of the SNPs within the known prostate cancer loci are located in the putative enhancer regions identified in human prostatic carcinoma cell (6). Indeed, enhancer-related dysregulation of gene expression has been recognized as one of the main drivers in the pathogenesis of many diseases (7,8). For example, in certain cases of cancer, the MYC oncogene is commonly translocated close to the enhancer regions (9,10).
Thanks to recent rapid development of sequencing technology, genome annotation consortia—e.g. ENCODE (11), FANTOM (12,13), NIH Epigenome Roadmap (14)—have generated a massive amount of the different types of sequencing data, which makes it possible for the identification of enhancers on the genome-wide scale. Although several databases have been set up for enhancers—e.g. DENDB (15) and EnhacerAtlas (16)—and super-enhancers—e.g. SEA (17) and dbSUPER (18)—in the human genome, they only provide limited basic information about enhancers, such as their coordinates, cell or tissue types, and nearby genes. As enhancers are highly relevant to human diseases, information about their disease connections can help us to better understand their potential roles in the biological processes of human diseases.
To facilitate studies of enhancers and their roles in the molecular mechanism of human complex diseases, we developed the Human Enhancer Disease Database (HEDD), the first integrated and interactive online knowledge base of enhancers and their disease associations. Compared with earlier released enhancer-related databases, HEDD contains the most up-to-date and complete set of enhancers in the human genome. It not only provides comprehensive genomic annotation on every enhancer in the database but also makes connections between enhancers and diseases and scores them using a newly developed scoring method. Moreover, HEDD offers Web-based analytical tools to visualize enhancer networks and score enhancers given a set of selected genes in a specific gene network. Overall, as a comprehensive enhancer resource with a focus on diseases, HEDD provides a convenient platform to search, browse, and download data related to enhancer and enhancer-disease association and facilitates studies of enhancers and their roles in human complex diseases.
MATERIALS AND METHODS
System design and implementation
The current version of HEDD has been developed using MySQL 5.7.17 (http://www.mysql.com) and runs on a Linux-based Apache Web server. PHP 5.4.16 (http://www.php.net/) is used for server-side scripting. We design and build the interactive interface using Bootstrap 3 (http://getbootstrap.com/), the most popular HTML, CSS and JS framework on the Web. We recommend using a modern Web browser such as Firefox (preferred), Google Chrome, or Safari to achieve the best display effect.
Data sources
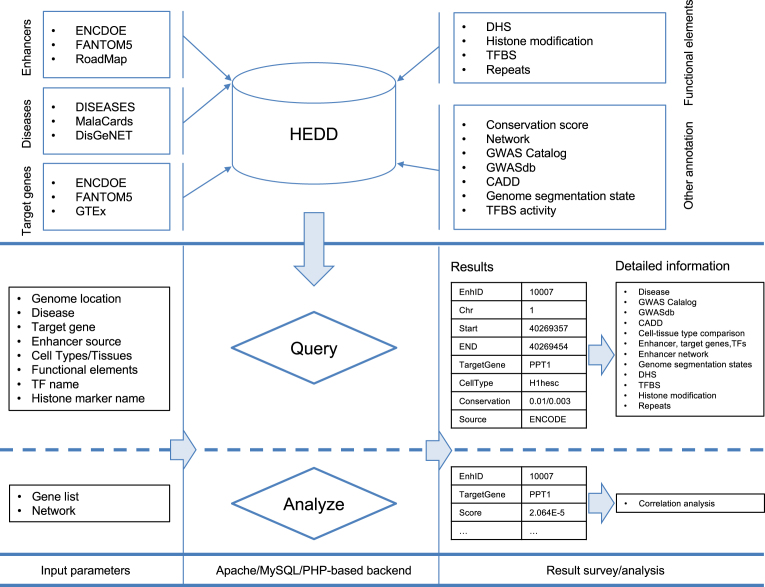
We integrated different sources of enhancers, human diseases, and functional genomic annotation to construct a central repository of human enhancers and their disease associations (Figure 1 and Table 1).
Figure 1.
Database content and construction. HEDD collected the enhancers (from three major epigenome study projects), enhancer target gene and gene disease set to quantify the connections between enhancer and diseases. Besides the disease information, it also stores the genetic and epigenetic information (e.g. DHS, TFBS, conservation score) related to enhancers. Users can query with multiple options (e.g. genome locations, disease name, gene name) to acquire enhancers and further view all the detail information such as, associated disease, cell type/tissue, overlapped GWAS SNPs, CADD score and neighboring enhancer for a specific enhancer. It enables users to do scoring analysis: for a gene set of interest, users can score enhancers in a gene network for their ‘relatedness’ to the gene set. All results of query and analysis can be downloaded for further analysis. DHS: DNase I hypersensitive sites, GWAS: genome-wide association studies, TF: transcription factor, TFBS: transcription factor biding sites, CADD: combined annotation dependent depletion.
Table 1. Summary of data sources (as of April 2017).
| Source | Cell-types/tissue | Number of records | Total | |
|---|---|---|---|---|
| Enhancer | ENCODE | 6 | 399 124 | 2 793 316 |
| FANTOM5 | — | 65 359 | ||
| RoadMap | 111 | 2 328 833 | ||
| Disease | DISEASES | — | 44 581 | 523 109 |
| MalaCards | — | 49 492 | ||
| DisGeNET | — | 429 036 | ||
| GWAS | GWAS Catalog | — | 35 329 | 349 566 |
| GWASdb v2 | — | 314 237 | ||
| SNV | CADD | — | ∼8.6 billion | ∼8.6 billion |
| Genome segmentation state | UCSC | 6 | 11 062 356 | 11 062 356 |
| DHS | UCSC | 120 | 10 040 306 | 10 040 306 |
| TFBS activity | Ensembl | 68 | 22 801 | 22 801 |
| TFBS | UCSC | 91 | 1611 | 1611 |
| Histone modification | UCSC | 19 | 411 | 411 |
| Repeats | UCSC | — | — | 1 533 636 |
| Conservation | UCSC | — | — | |
| Target gene connection | ENCODE | — | 13 812 | |
| FANTOM5 | 66 943 | |||
| GTEx | 26 393 329 | |||
| Network | Nodes | Edges | ||
| HINT | 11 984 | 53 405 | ||
| HPRD | 9 460 | 36 985 | ||
| HIPPIE | 16 567 | 276 051 | ||
| PIPs | 5 445 | 37 343 | ||
| CCSB | 4 230 | 13 427 | ||
| IID | 18 080 | 915 091 | ||
| UniHI | 17 685 | 364 777 |
Note: 1. The number of markers.
Enhancers
The current release of HEDD makes available a total of 2 793 316 putative enhancers collected from three major genome/epigenome annotation projects: 399 124 from ENCODE (11) predicted jointly by two segmentation methods—ChromHMM and Segway (19), 65 359 from FANTOM5 (12,13) predicted by the cap analysis of gene expression (CAGE) (20) and 2 328 833 from RoadMap (14) predicted by ChromHMM (21).
Functional genomic annotation of enhancers
The genomic regions of enhancers usually have several prominent features, including DNase I hypersensitivity, transcription factor biding sites (TFBS), and enriched histone acetylation (22–24). To build an enhancer knowledge base, we collected six types of functional genomic annotation: (i) DNase I hypersensitive sites (DHS) (25,26), (ii) transcription factor binding sites (26,27), (iii) histone modification marks (26), (iv) repeats (28), (v) genome segmentation states (11), and (vi) evolutionary conservation (28). See Table 1 for a summary of these data sets.
Enhancer target genes
To study the biological function and the disease association of enhancers, it is critical to annotate their target genes. We collected enhancer target genes from three sources: (i) the genome-wide map of distal DHS-to-promoter connectivity data from ENCODE (11), (ii) intra-chromosomal enhancer-promoter expression correlation data from FANTOM (12) and (iii) eQTL and target gene data from GTEx (29) for enhancers identified by the RoadMap Project.
Human diseases
We collected the human genes-disease association data from MalaCards (30), DISEASES (31) and DisGeNET (32). These databases provide both genes–disease pairs and the scores representing the strength of association between them. In addition, we annotated enhancers with disease/traits-associated or deleterious genetic variants from GWAS Catalog, GWASdb (33,34) and CADD (35), which provides scores of functional deleteriousness for both single nucleotide variants and insertion/deletions variants in the human genome.
Gene networks
As part of HEDD, we provide seven gene networks—HINT (High-quality Interactomes) (36), HPRD (Human Protein Reference Database) (37), HIPPIE (Human Integrated Protein Protein Interaction Reference) (38), PIPs (protein–protein interactions) (39), CCSB (Center for Cancer Systems Biology) (40), IID (Integrated Interactions Database) (41), UniHI (Unified Human Interactome) (42)—which are used to score enhancers in the context of the a biological network.
Evolutionary conservation of genomic sequences
We quantified the evolutionary conservation of enhancers among placental mammals using the sequence conservation scores for positions in the human genome from the UCSC genome browser (28). HEDD provides summary statistics (the mean and the median) of conservation scores for enhancers.
Score the enhancer-disease connection
To study enhancer-related disease mechanisms, the first major challenge is to confidently link enhancers to diseases. We used disease-associated genes as the intermediaries to build such connections: if an enhancer targets a disease-associated gene, then this enhancer is functionally connected to the disease. As connections between enhancers and genes and between genes and diseases are both scored and their scores have the same directionality (the higher the score, the stronger the connection), we were able to quantify the connections between enhancers and diseases. To do this, we first calculated the percentile for every score in a set of scores from a particular source as the probability of connection between an enhancer and a gene (pEG) or between a gene and a disease (pGD). We then computed the probability of connection between an enhancer and a disease (pED) by multiplying pEG and pGD, the two probabilities of their respective connections with an intermediate gene: pED = pEG × pGD. In this way, we connect enhancers and diseases through genes that are connected to both of them, and also quantify the strength of their connections.
Score enhancers based on a gene set
Given a gene set of interest (e.g. differentially expressed genes or disease/trait-related genes), an online software tool in HEDD can score enhancers based on their connections to genes and the centrality of genes in a gene network using the highly successful Google PageRank algorithm (43) that we implemented before (44). Briefly, for a set of genes, HEDD first scores the relatedness of each gene in a gene network to the gene set based on how all genes are wired in the network to the gene set. These scores are then transferred from genes to their enhancers. The score indicating the strength of connection between an enhancer and the gene set is defined by the mean or the sum of scores from all the target genes of this enhancer.
Gene set-disease correlation analysis
Given a functionally coherent set of genes (e.g. from a differential gene expression analysis), HEDD can suggest their related diseases as a form of functional annotation based on the correlation between two sets of enhancer scores: scores of connections between enhancers and a disease and scores of enhancers based on the gene set in the gene network. With the first set of enhancer scores pre-computed for a number of diseases (and stored in HEDD), this correlation analysis uses enhancers as intermediates to link and score the connections between a gene set and diseases. The disease with the highest correlation coefficient could shed light on the molecular function and biological process of the given gene set.
DATABASE USE AND ACCESS
Database search and browsing
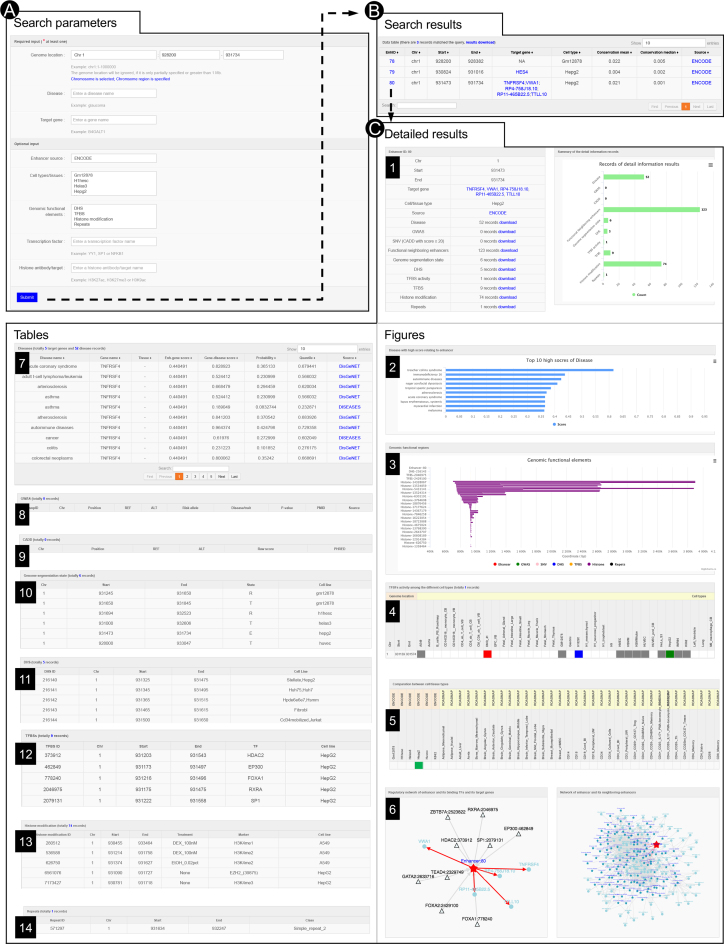
HEDD can be interactively searched and browsed in various ways (Figure 2). Users can search for enhancers in a genomic region, disease, target gene, or by conditions such as source, cell type/tissue, overlapped functional element (DHS, TFBS, histone mark, and repeat), transcription factor, and histone modification marker (Figure 2A). The search result, including genomic coordinates, target genes, cell type/tissues, conservation scores, and sources, are organized and returned in a tabular format (Figure 2B). Following links embedded in the enhancer IDs, users can examine on a new webpage detailed annotation of every enhancer in the table (Figure 2), including information about its related diseases (Figure 2C2 and 2C7) with corresponding scores (and their quantiles), functional annotation of its variants from GWAS and CADD (Figure 2C8–9), cell/tissue types, enhancer network with its target genes, TFBS and neighboring enhancers, genome segmentation states, TFBS activity, and overlapped genomic elements (e.g. DHS, TFBS, histone modification and repeats; Figure 2C3–6 and 2C10–14). All search results can be downloaded (Figure 2C1) for further analysis.
Figure 2.
Interactive searching and browsing activity of HEDD. (A) Input parameters for query. (B) The result table, including enhancer IDs, genomic coordinates, target genes, cell types/tissues, conservation scores and sources. (C) Details of a selected enhancer from the result table, including its associated diseases, functional annotation from GWAS and CADD, overlapped genomic elements such as DHS, TFBSs, histone modification and repeats, TFBSs activity levels, genome segmentation states, the comparison among cell/tissues types, regulatory network, and neighboring enhancers.
Online analysis tools
On the ‘Analyze’ webpage, for a gene set of interest, users can score enhancers in a gene network for their ‘relatedness’ to the gene set (Supplementary Figure S1). User can either select a gene network out of seven that are currently available or upload a customer network (Supplementary Figure S1A). The scores (both mean and sum) are returned in a descending order in a table (Supplementary Figure S1B2). Input genes that are present or absent in the selected/uploaded gene network will also be reported (Supplementary Figure S1B1). Users can use these scores in a subsequent correlation analysis (Supplementary Figure S1C), which diseases with highest absolute correlation coefficients (top 20) will be shown in bar charts (Supplementary Figure S1D). We used a list of 242 schizophrenia genes to benchmark the running time for all networks that we make available online. It usually takes several minutes to score enhancers, depending on the selected gene network, and correlation analysis takes about half an hour.
APPLICATION
Analysis of enhancer distribution and disease association in 9p21 locus
Chromosome 9p21 locus is a 13.3-Mb gene-poor genomic region, which contains many genetic variants associated with multiple human complex diseases, including coronary artery disease (CAD), glaucoma, diabetes, and several cancers. Most of the risk variants in this region are non-coding, suggesting that they influence gene expression and may act in cis (45). We analyzed the genomic distribution of enhancers in 9p21 and found eight regions with high densities of enhancers (Supplementary Figure S3A, Supplementary Table S1). The genomic distribution of enhancers with disease association is mostly consistent with the overall distribution of enhancers. Using enhancer-disease associations with the top 10% highest scores, we screened for diseases associated with each enhancer cluster and found the majority of these clusters are highly associated with cancer (Supplementary Figure S3B, Supplementary Table S1).
Enhancers overlapping or near these risk variants could be the genomic functional elements underlying their disease association. Indeed, we identified several blocks of 9p21 region in which enhancers are scored high for corresponding variant-associated diseases (Supplementary Figure S2A). Block B contains two genomic regions enriched with risk SNPs of glaucoma and CAD (Supplementary Figure S2B). We found that enhancers in high linkage disequilibrium with those risk SNPs—rs523096 (46), rs4977756 (47,48), rs1333037 (49), rs1063192 (50), rs7865618 (51), rs2157719 (52) and rs7866783 (53) for glaucoma; rs1537370 (54), rs1333049 (55,56), rs10738607 (57), rs4977574 (58) and rs2891168 (59) for CAD—have the highest scores with these two diseases in this block. In block C, all the enhancers have the highest score with the small cell lung cancer (SCLC) among other diseases. Near one of these enhancers is a SNP—rs4246856—associated with D-dimer level (Supplementary Figure S2C), which has been shown to provide useful information for predicting the prognosis of patients with SCLC (60). Block D contains enhancers in TEK, a gene that also contains a SNP (rs2273720) associated with endothelial growth factor levels that are correlated with the formation of blood vessels. Interestingly, enhancers in this block have the highest scores with venous malformations, multiple cutaneous and mucosal, diseases related to blood vessels (Supplementary Figure S2D). Block E and F contains genes with multiple risk SNPs associated with obesity (Supplementary Figure S2E) and amyotrophic lateral sclerosis (Supplementary Figure S2F), respectively. Enhancers of these genes show higher scores for those two diseases than other diseases. In block G, an enhancer strongly associated with congenital disorder of glycosylation was found near rs10971170 (61), a risk SNP related to igG glycosylation (Supplementary Figure S2G), and could be the regulatory element underlying the genetic signal of the risk SNP.
Identification of potential regulatory causal variants for human complex diseases
HEDD can be used to identify potential regulatory causal variants in enhancers in post-GWAS analyses of human complex diseases. We analyzed glaucoma GWAS results as an example of this usage. We first collected from the GWAS Catalog (33) 51 glaucoma-associated SNPs. To use linkage disequilibrium (LD) as mapping tool to find potential casual variants in enhancer, we searched their vicinity (within 25 kb upstream and downstream) and found near 36 of them 2871 enhancers containing 42 923 variants in total (based on NCBI dbSNP as of April 2017). 907 enhancer variants have alleles with high CADD scores (≥ 20) and thus were considered as candidates of causal variants. For 129 of them, here is genotype information (from 2,504 individuals) from the 1000 Genomes Project. We calculated the LD between one of the 36 GWAS SNPs and one of the 129 causal variant candidates in the former's neighborhood using the 1000 Genomes Project (Phase 3) genotype data. We identified one potential regulatory causal variant (rs8940). This enhancer variant is in relatively high linkage disequilibrium (r2 = 0.565) with the glaucoma-associated SNP rs4236601. Interestingly, a previous study has reported that SNP rs8940 was associated with glaucoma (62).
Building disease gene regulatory networks
Human complex diseases are results of gene dysregulation. It is thus particularly important to elucidate the regulatory relationship among genes related to a complex disease. Using enhancer-disease association data in HEDD, we can build a gene regulatory network for a complex disease based on enhancers associated with it and their connections with transcription factors and target genes. Using acute erythroblastic leukemia (AEL) as a disease example, we found 651 enhancers with high AEL-association scores (≥ 0.6). These enhancers are connected to 37 genes, including seven transcription factor genes. We built the gene regulatory network for AEL based on these 37 genes (five genes without the interaction with other genes were removed, Supplementary Figure S3C). Transcriptional factor genes—GATA1, TAL1, SPI1, STAT1, STAT3—display high network centrality, implying their important roles in the regulatory mechanism related to the disease pathology. Few of interactions in the network have already been reported previously, except for TAL1 targeting GATA1 (63,64) and GYPA (65). Therefore, gene regulatory network built with HEDD enhancer data can be used to generate hypothesis about regulatory mechanisms of disease pathology.
CONCLUSION AND FUTURE DEVELOPMENT
We have built an integrated database for human enhancers and their disease associations. Our goal is to provide a comprehensive data resource and a set of interactive analysis tools to facilitate genomic research of enhancers and their roles in human complex diseases. We will continue to update the database with the latest data sets when they become available. In the future, we will add more genetic and epigenetic information about enhancers, such as the topologic associated domains and the retargeting of enhancers in different cell type/tissues or cancers. We believe that our enhancer database will be of particular interest to researchers working on the gene regulatory networks of human diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Yousin Suh and Michael G. Rosenfeld for their expert advice on the biology of enhancers.
Authors’ contributions: Z.D.Z. conceived and designed this study. Z.W. carried out the analyses and also participated in the study design. Z.W. and Z.D.Z. wrote the manuscript. Q.Z., J.R.L., Y.C. and J.M. provided useful input for the analyses and helped edit the manuscript. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 HG008153] from the National Human Genome Research Institute (to Z.D.Z.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Corradin O., Scacheri P.C.. Enhancer variants: evaluating functions in common disease. Genome Med. 2014; 6:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J. et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farh K.K.H., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J.H., Shishkin A.A. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015; 518:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gjoneska E., Pfenning A.R., Mathys H., Quon G., Kundaje A., Tsai L.H., Kellis M.. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature. 2015; 518:365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hazelett D.J., Rhie S.K., Gaddis M., Yan C.L., Lakeland D.L., Coetzee S.G., Henderson B.E., Noushmehr H., Cozen W., Kote-Jarai Z. et al. Comprehensive functional annotation of 77 prostate cancer risk loci. Plos Genet. 2014; 10, doi:10.1371/journal.pgen.1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sur I., Taipale J.. The role of enhancers in cancer. Nat. Rev. Cancer. 2016; 16:483–493. [DOI] [PubMed] [Google Scholar]
- 8. Herz H.M. Enhancer deregulation in cancer and other diseases. Bioessays. 2016; 38:1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. ar-Rushdi A., Nishikura K., Erikson J., Watt R., Rovera G., Croce C.M.. Differential expression of the translocated and the untranslocated c-myc oncogene in Burkitt lymphoma. Science. 1983; 222:390–393. [DOI] [PubMed] [Google Scholar]
- 10. Erikson J., ar-Rushdi A., Drwinga H.L., Nowell P.C., Croce C.M.. Transcriptional activation of the translocated c-myc oncogene in burkitt lymphoma. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:820–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R. et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forrest A.R.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J.L., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M. et al. A promoter-level mammalian expression atlas. Nature. 2014; 507:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. et al. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010; 28:1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ashoor H., Kleftogiannis D., Radovanovic A., Bajic V.B.. DENdb: database of integrated human enhancers. Database-Oxford. 2015; 2015:bav085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao T., He B., Liu S., Zhu H., Tan K., Qian J.. EnhancerAtlas: a resource for enhancer annotation and analysis in 105 human cell/tissue types. Bioinformatics. 2016; 32:3543–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wei Y.J., Zhang S.M., Shang S.P., Zhang B., Li S., Wang X.Y., Wang F., Su J.Z., Wu Q., Liu H.B. et al. SEA: a super-enhancer archive. Nucleic Acids Res. 2016; 44:D172–D179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khan A., Zhang X.G.. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016; 44:D164–D171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoffman M.M., Ernst J., Wilder S.P., Kundaje A., Harris R.S., Libbrecht M., Giardine B., Ellenbogen P.M., Bilmes J.A., Birney E. et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013; 41:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M. et al. CAGE: cap analysis of gene expression. Nat. Methods. 2006; 3:211–222. [DOI] [PubMed] [Google Scholar]
- 21. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W.B., Notani D., Rosenfeld M.G.. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 2016; 17:207–223. [DOI] [PubMed] [Google Scholar]
- 23. Bulger M., Groudine M.. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011; 144:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blackwood E.M., Kadonaga J.T.. Going the distance: a current view of enhancer action. Science. 1998; 281:60–63. [DOI] [PubMed] [Google Scholar]
- 25. Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. et al. The accessible chromatin landscape of the human genome. Nature. 2012; 489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G. et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013; 41:D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zerbino D.R., Wilder S.P., Johnson N., Juettemann T., Flicek P.R.. The ensembl regulatory build. Genome Biol. 2015; 16:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M. et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015; 43:D670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N. et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rappaport N., Nativ N., Stelzer G., Twik M., Guan-Golan Y., Stein T.I., Bahir I., Belinky F., Morrey C.P., Safran M. et al. MalaCards: an integrated compendium for diseases and their annotation. Database-Oxford. 2013; 2013:bat018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pletscher-Frankild S., Palleja A., Tsafou K., Binder J.X., Jensen L.J.. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015; 74:83–89. [DOI] [PubMed] [Google Scholar]
- 32. Pinero J., Queralt-Rosinach N., Bravo A., Deu-Pons J., Bauer-Mehren A., Baron M., Sanz F., Furlong L.I.. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database-Oxford. 2015; 2015:bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014; 42:D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li M.J., Liu Z.P., Wang P.W., Wong M.P., Nelson M.R., Kocher J.P.A., Yeager M., Sham P.C., Chanock S.J., Xia Z.Y. et al. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016; 44:D869–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Das J., Yu H.Y.. HINT: High-quality protein interactomes and their applications in understanding human disease. BMC Syst. Biol. 2012; 6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prasad T.S.K., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A. et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009; 37:D767–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alanis-Lobato G., Andrade-Navarro M.A., Schaefer M.H.. HIPPIE v2.0: enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res. 2017; 45:D408–D414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDowall M.D., Scott M.S., Barton G.J.. PIPs: human protein-protein interaction prediction database. Nucleic Acids Res. 2009; 37:D651–D656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rolland T., Tasan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R. et al. A proteome-scale map of the human interactome network. Cell. 2014; 159:1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kotlyar M., Pastrello C., Sheahan N., Jurisica I.. Integrated interactions database: tissue-specific view of the human and model organism interactomes. Nucleic Acids Res. 2016; 44:D536–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kalathur R.K.R., Pinto J.P., Hernandez-Prieto M.A., Machado R.S.R., Almeida D., Chaurasia G., Futschik M.E.. UniHI 7: an enhanced database for retrieval and interactive analysis of human molecular interaction networks. Nucleic Acids Res. 2014; 42:D408–D414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Page L., Brin S., Motwani R., Winograd T.. The PageRank Citation Ranking: Bringing Order to the Web. 1999; Stanford InfoLab, Technical Report. [Google Scholar]
- 44. Lemetrey C., Zhang Q.W., Zhang Z.D.D.. SubNet: a Java application for subnetwork extraction (vol 29, pg 2509, 2013). Bioinformatics. 2013; 29:2958–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cunnington M.S., Santibanez Koref M., Mayosi B.M., Burn J., Keavney B.. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL Expression. PLoS Genet. 2010; 6:e1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takamoto M., Kaburaki T., Mabuchi A., Araie M., Amano S., Aihara M., Tomidokoro A., Iwase A., Mabuchi F., Kashiwagi K. et al. Common variants on chromosome 9p21 are associated with normal tension glaucoma. PLoS One. 2012; 7:e40107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burdon K.P., Macgregor S., Hewitt A.W., Sharma S., Chidlow G., Mills R.A., Danoy P., Casson R., Viswanathan A.C., Liu J.Z. et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat. Genet. 2011; 43:574–578. [DOI] [PubMed] [Google Scholar]
- 48. Gharahkhani P., Burdon K.P., Fogarty R., Sharma S., Hewitt A.W., Martin S., Law M.H., Cremin K., Bailey J.N., Loomis S.J. et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat. Genet. 2014; 46:1120–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bailey J.N., Loomis S.J., Kang J.H., Allingham R.R., Gharahkhani P., Khor C.C., Burdon K.P., Aschard H., Chasman D.I., Igo R.P. Jr et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat. Genet. 2016; 48:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Osman W., Low S.K., Takahashi A., Kubo M., Nakamura Y.. A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum. Mol. Genet. 2012; 21:2836–2842. [DOI] [PubMed] [Google Scholar]
- 51. Nakano M., Ikeda Y., Tokuda Y., Fuwa M., Omi N., Ueno M., Imai K., Adachi H., Kageyama M., Mori K. et al. Common variants in CDKN2B-AS1 associated with optic-nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PLoS One. 2012; 7:e33389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wiggs J.L., Yaspan B.L., Hauser M.A., Kang J.H., Allingham R.R., Olson L.M., Abdrabou W., Fan B.J., Wang D.Y., Brodeur W. et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. Plos Genet. 2012; 8:e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Verma S.S., Cooke Bailey J.N., Lucas A., Bradford Y., Linneman J.G., Hauser M.A., Pasquale L.R., Peissig P.L., Brilliant M.H., McCarty C.A. et al. Epistatic gene-based interaction analyses for glaucoma in eMERGE and NEIGHBOR consortium. PLoS Genet. 2016; 12:e1006186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van Setten J., Isgum I., Smolonska J., Ripke S., de Jong P.A., Oudkerk M., de Koning H., Lammers J.W., Zanen P., Groen H.J. et al. Genome-wide association study of coronary and aortic calcification implicates risk loci for coronary artery disease and myocardial infarction. Atherosclerosis. 2013; 228:400–405. [DOI] [PubMed] [Google Scholar]
- 55. O’Donnell C.J., Kavousi M., Smith A.V., Kardia S.L., Feitosa M.F., Hwang S.J., Sun Y.V., Province M.A., Aspelund T., Dehghan A. et al. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation. 2011; 124:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dichgans M., Malik R., Konig I.R., Rosand J., Clarke R., Gretarsdottir S., Thorleifsson G., Mitchell B.D., Assimes T.L., Levi C. et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke. 2014; 45:24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wakil S.M., Ram R., Muiya N.P., Mehta M., Andres E., Mazhar N., Baz B., Hagos S., Alshahid M., Meyer B.F. et al. A genome-wide association study reveals susceptibility loci for myocardial infarction/coronary artery disease in Saudi Arabs. Atherosclerosis. 2016; 245:62–70. [DOI] [PubMed] [Google Scholar]
- 58. Reilly M.P., Li M., He J., Ferguson J.F., Stylianou I.M., Mehta N.N., Burnett M.S., Devaney J.M., Knouff C.W., Thompson J.R. et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011; 377:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C. et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015; 47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smith N.L., Huffman J.E., Strachan D.P., Huang J., Dehghan A., Trompet S., Lopez L.M., Shin S.Y., Baumert J., Vitart V. et al. Genetic predictors of fibrin D-dimer levels in healthy adults. Circulation. 2011; 123:1864–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lauc G., Huffman J.E., Pucic M., Zgaga L., Adamczyk B., Muzinic A., Novokmet M., Polasek O., Gornik O., Kristic J. et al. Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet. 2013; 9:e1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Williams S.E.I. The Genetics of Primary Open-angle Glaucoma in Black South Africans (Doctoral dissertation). 2014; University of the Witwatersrand, Faculty of Health Sciences. [Google Scholar]
- 63. Vyas P., McDevitt M.A., Cantor A.B., Katz S.G., Fujiwara Y., Orkin S.H.. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development. 1999; 126:2799–2811. [DOI] [PubMed] [Google Scholar]
- 64. Kassouf M.T., Hughes J.R., Taylor S., McGowan S.J., Soneji S., Green A.L., Vyas P., Porcher C.. Genome-wide identification of TAL1’s functional targets: Insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010; 20:1064–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lahlil R., Lecuyer E., Herblot S., Hoang T.. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol. Cell. Biol. 2004; 24:1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.