Abstract
Here, we described the updated database iUUCD 2.0 (http://iuucd.biocuckoo.org/) for ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), ubiquitin-protein ligases (E3s), deubiquitinating enzymes (DUBs), ubiquitin/ubiquitin-like binding domains (UBDs) and ubiquitin-like domains (ULDs), which act as key regulators in modulating ubiquitin and ubiquitin-like (UB/UBL) conjugations. In total, iUUCD 2.0 contained 136 512 UB/UBL regulators, including 1230 E1s, 5636 E2s, 93 343 E3s, 9548 DUBs, 30 173 UBDs and 11 099 ULDs in 148 eukaryotic species. In particular, we provided rich annotations for regulators of eight model organisms, especially in humans, by compiling and integrating the knowledge from nearly 70 widely used public databases that cover cancer mutations, single nucleotide polymorphisms (SNPs), mRNA expression, DNA and RNA elements, protein–protein interactions, protein 3D structures, disease-associated information, drug-target relations, post-translational modifications, DNA methylation and protein expression/proteomics. Compared with our previously developed UUCD 1.0 (∼0.41 GB), iUUCD 2.0 has a size of ∼32.1 GB of data with a >75-fold increase in data volume. We anticipate that iUUCD 2.0 can be a more useful resource for further study of UB/UBL conjugations.
INTRODUCTION
Ubiquitin and ubiquitin-like (UB/UBL) conjugations are widespread regulatory post-translational modifications (PTMs) of proteins; they regulate a broad spectrum of cellular processes such as protein degradation and turnover, intercellular signal transduction, cell cycle and DNA damage repair (1–3). The dysregulation of protein ubiquitination and cognate PTMs is highly involved in a number of human pathologies such as tumorigenesis, neurodegeneration and cardiac diseases (4,5). In this regard, the identification of UB/UBL regulators and substrate proteins is fundamental for understanding the molecular mechanisms and functional roles of UB/UBL conjugation and provides potential targets for further drug design (6).
Biochemically, UB/UBL conjugations are catalyzed through a similar three-step thioester cascade process sequentially performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) (7), while the modifications can be reversed by deubiquitinating enzymes (DUBs) to remove UB/UBL moieties from protein substrates (8). In addition to these enzymes and adaptors, numerous proteins with UB/UBL binding domains (UBDs) or ubiquitin-like domains (ULDs, also known as UBQs, ubiquitin homologs) also play essential roles in the regulation of UB/UBL conjugation (8,9). In general, the specificity of ubiquitin signaling is conferred by alternative conjugation types and interactions with UBDs that selectively recognize monoubiquitin and polyubiquitin chains with different linkages and lengths to control various cellular functions in vivo (10,11). ULD-containing proteins can be classified as UBL modifiers and ubiquitin domain proteins (UDPs) (12–14). Both UBLs and UDPs have limited sequence similarity with ubiquitin but exhibit a conserved globular β-grasp ubiquitin superfold, whereas UDPs contain one or multiple integral ULDs in the protein sequence (12–14). In particular, a considerable number of proteins contain both UBD and ULD domains, and these proteins mainly act as ‘shuttle buses’ to target polyubiquitinated proteins to the proteasome for degradation (14).
With the growing number of identified UB/UBL regulators, the collection, classification and integration of these proteins will provide a useful resource for further research. In 2013, we developed a family-based database, UUCD 1.0, containing 738 E1s, 2937 E2s, 46,631 E3s and 6647 DUBs in 70 eukaryotic organisms (15). Almost at the same time, Hutchins et al. also assembled a similar database, DUDE-db, which collected and identified 35 228 unique ubiquitinating enzymes (UBEs) and DUBs from 50 genomes (16). These authors also performed multiple phylogenetic analyses of UBEs and DUBs for a better understanding of the evolutionary history of the ubiquitination system in eukaryotes (16). However, UBD and ULD proteins were not included in either database. In 2016, Harrison et al. first constructed a highly useful database, UbSRD, that contained 509 UBL-containing protein 3D structures characterized from the Protein Data Bank (PDB) (17) as well as a platform for the structural analysis (18). However, a systematic identification of UBD and ULD proteins in eukaryotes remains to be carried out.
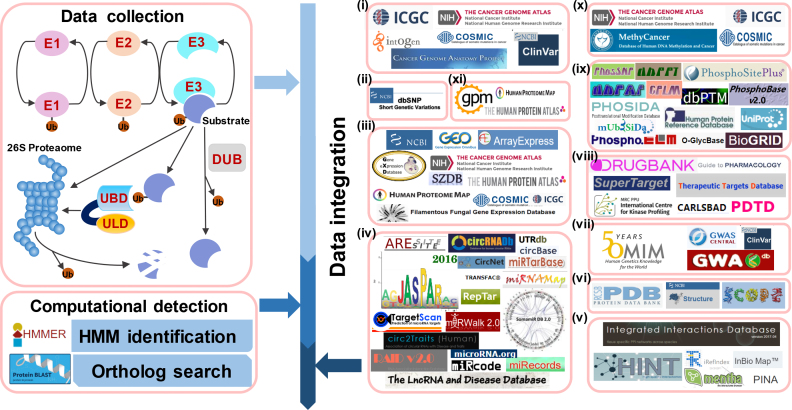
In this update, we greatly improved our previous UUCD 1.0 database and provided a much more comprehensive resource, namely, iUUCD 2.0. From the literature, we focused on experimentally identified UB/UBL regulators; we collected 27 E1s, 109 E2s, 1153 E3s, 164 DUBs, 396 UBDs and 183 ULDs (Figure 1). Based on the consensus of previous studies (10,11,14,15,19), we classified known regulators into 1, 4, 23, 8, 27 and 11 families. For families with ≥3 genes, we used HMMER (20) and obtained 1, 2, 17, 7, 22 and 9 hidden Markov model (HMM) profiles for E1s, E2s, E3s, DUBs, UBDs and ULDs at the family level, respectively. Next, we used these HMM profiles to conduct a genome-wide identification and systematically identified 978 E1s, 5053 E2s, 71 907 E3s, 7379 DUBs, 22 330 UBDs and 8464 ULDs in 148 eukaryotes (Figure 1). For families without HMMs, we also used the known regulators to perform an additional orthologous search to verify the data integrity (Figure 1). In addition to a number of basic annotations such as accession numbers, gene/protein names, protein/nucleotide sequences, domain/motifs, active sites of UB/UBL enzymes, functional descriptions and gene ontology (GO) terms obtained from Ensembl (21) and UniProt (22) databases, we further mapped all identified human regulators to 67 additional public resources to obtain rich annotations. We also mapped these annotations to regulators in seven additional model organisms if available. In total, iUUCD 2.0 contained 136 512 UB/UBL regulators (1230 E1s, 5636 E2s, 93 343 E3s, 9548 DUBs, 30 173 UBDs and 11 099 ULDs) in 148 eukaryotic species with a data volume of ∼32.1 GB. The iUUCD database is free for all users at: http://iuucd.biocuckoo.org/.
Figure 1.
The procedure for the construction of the iUUCD 2.0 database. First, we searched PubMed to collect experimentally identified UB/UBL regulators. Then, we classified known regulators into distinct families and constructed HMM profiles for each family if available. For families without HMM profiles, we also conducted an orthologous search to ensure data integrity. In addition to basic annotations, we further mapped all regulators in eight model organisms, especially in Homo sapiens, to nearly 70 public databases that covered each of 11 aspects: (i) cancer mutations, (ii) SNPs, (iii) mRNA expressions, (iv) DNA and RNA elements, (v) PPIs, (vi) protein 3D structures, (vii) disease-associated information, (viii) drug-target relations, (ix) PTMs, (x) DNA methylation and (xi) protein expression/proteomics.
CONSTRUCTION AND CONTENT
Data collection
In UUCD 1.0, we collected 26 E1s, 105 E2s, 1003 E3s and 148 DUBs from the literature (15). Here, we mainly focused on the biocuration of newly reported enzymes and adaptors published since 2013. To collect known UBD- and ULD-containing proteins, we first searched the PubMed database using multiple general keywords such as ‘ubiquitin binding’, ‘ubiquitin-binding domain’, ‘ubiquitin-like protein’ and ‘ubiquitin-like domain’. Then, all related papers were downloaded with a careful curation. To avoid missing any important regulators, we used each family name to search the PubMed again after the classification of known UB/UBL regulators. In total, we obtained 27 E1s, 109 E2s, 1153 E3s, 164 DUBs, 396 UBD proteins and 183 ULD proteins (Supplementary Table S1).
For the known regulators, we retrieved their protein sequences from the Ensembl (21) and UniProt (22) databases. The functional domain information was acquired and further confirmed by searching the UniProt (22), Pfam (23) and InterPro databases (24). For the large-scale identification, the complete proteome sequences were downloaded for 148 eukaryotes (68 animals from Ensembl (release version 87, http://www.ensembl.org/), 39 plants from EnsemblPlants (release version 34, http://plants.ensembl.org/) and 41 fungi from EnsemblFungi (release version 34, http://fungi.ensembl.org/)). As previously described (15), multiple variant nucleotide/protein sequences can be derived from a single gene, and here we adopted the Ensembl Gene ID as the unique accession to avoid redundancy. For multiple alternative splicing isoforms of a gene, only the longest form was included.
The classification of UB/UBL regulators
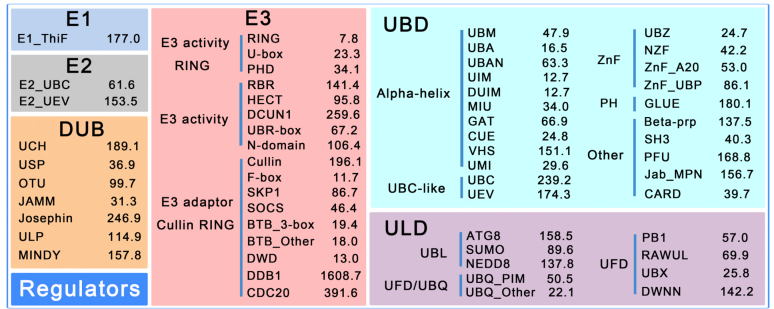
Previously we classified known E1s, E2s, E3s and DUBs into 1, 3, 19 and 7 families, respectively (15). Due to the new data accumulation, we re-checked the classifications and added 6 new families for E2s, E3s and DUBs (19,25–29): Autophagy_C (Autophagy-related C-terminal), PHD (plant homeodomain finger), A20 (an inhibitor of cell death), DCUN1 (defective in cullin neddylation), RBR (RING-between RING-RING) and MINDY (motif interacting with Ub-containing novel DUB family). The Autophagy_C family belongs to E2s, PHD, A20, DCUN1 and RBR are in the E3 family, whereas MINDY belongs to DUBs (Figure 2).
Figure 2.
The classification of UB/UBL regulators together with the cut-off values of the 58 HMM profiles for E1, E2, E3, DUB, UBD and ULD families. The hmmsearch program calculates both the E-values and log-odds likelihood scores for given protein sequences. Since the E-values depend on the database size and generate inconsistent results when the database is updated, we used realistic constant log-odds likelihood scores as the threshold values.
Based on the consensus reached from various experimental studies (10,11,14), a hierarchical classification of UBD and ULD families was established in iUUCD 2.0 (Figure 2). First, we classified all UBD proteins into six groups: alpha-helix (α-helical-based structures), ZnF (zinc-finger), UBC-like (ubiquitin-conjugating-like), PH (pleckstrin homology), SIM (SUMO-interacting motif) and Other (10,30). Because a majority of UBDs fold into α-helical-based structures, the alpha-helix group consisted of 11 families (10): UBA (ubiquitin-associated), UBM (ubiquitin-binding motif), UBAN (UBD in ABIN proteins and NEMO), CUE (coupling of ubiquitin conjugation to endoplasmic reticulum degradation), GAT [GGA (Golgi-localized, gamma-ear-containing, ADP-ribosylation-factor-binding protein) and TOM (target of Myb)], MIU (motif interacting with ubiquitin), UIM (ubiquitin-interacting motif), DUIM (double-sided ubiquitin-interacting motif), VHS [Vps (vacuolar sorting protein) 27/h/STAM], UMI (UIM-and-MIU-related) and WIYLD (a three-alpha helices containing domain). The ZnF group contains five families: NZF (Npl4 ZnF), UBZ (ubiquitin-binding ZnF), ZnF_A20 (A20-type ZnF), ZnF_UBP (UBP-type ZnF) and ZnF/Other. Moreover, the UBC-like group was classified into two families [UBC (ubiquitin-conjugating) and UEV (UBC E2 variant)], whereas the PH group had only one family, GLUE (GRAM-like ubiquitin binding in EAP45). Additionally, the SIM group had only one family, SIM. In addition, proteins not included in the above five groups were classified into the Other group, which contains seven families: Beta-Prp (WD40-repeat β-propellers), CARD (Caspase recruitment domain), Jab_MPN (domain in Jun kinase and Mpr1p and Pad1p N-termini), PFU (PLAA family ubiquitin binding), PRU (pleckstrin-like receptor for ubiquitin), SH3 (SRC homology-3) and Other (unclassified proteins).
The classification of ULDs was relatively simple due to their structural similarity with ubiquitin (12–14). First, we classified all ULD proteins into two groups: UBL and UDP (12–14). The UBL group consisted of four families: SUMO (small ubiquitin-like modifier), ATG8 (ubiquitin-like protein Atg8), ATG12 (ubiquitin-like protein Atg12) and NEDD8 (neddylation domain). The UDP group, which usually bears an integral ULD at its N-terminal end, contained two subgroups, UBQ (ubiquitin homologs) and UFD (ubiquitin fold domain). The UBQ consisted of two families, UBQ_PIM (proteasome-interacting motif) and UBQ_Other, whereas UFD proteins were classified into five families (DWNN (domain with no name), PB1 (Phox and Bem1), RAWUL (RING finger- and WD40-associated ubiquitin-like), UBX (domain present in ubiquitin-regulatory proteins) and MUB (membrane ubiquitin).
The genome-wide identification
As previously described (15), the sequences of protein functional domains were first retrieved and multi-aligned by MUSCLE (http://www.drive5.com/muscle/, version 3.8.31) (31) for each family with ≥3 genes. Then, the hmmbuild program in the HMMER v3.1b2 package (http://hmmer.org/) (20) was used for the construction of an HMM profile for each family. By this approach, we constructed 1, 2, 17, 7, 22 and 9 HMM profiles for E1, E2, E3, DUB, UBD and ULD families, respectively. With these HMM profiles, we further searched all protein sequences in 148 eukaryotic species by using the hmmsearch program in HMMER (20). All HMM profiles can be downloaded at: http://iuucd.biocuckoo.org/download.php.
To evaluate the accuracy of HMM-based identification and determine the threshold for each family, we regarded the known proteins annotated in each family as positive data (P) and known proteins classified in other families as negative data (N). For each HMM profile, the sensitivity (Sn) and specificity (Sp) values were calculated as below:
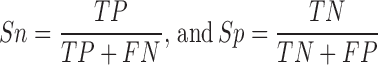 |
Both the self-consistency and leave-one-out (LOO) validations were performed. The receiver operating characteristic (ROC) curves were drawn, and AUC (area under ROC) values were computed for each family. For simplicity, the results of nine families are shown; obviously the predictions were accurate and robust (Supplementary Figure S1). To verify that all curated proteins were correctly predicted and classified (Sn = 100%), we manually selected the optimal cut-off value for each family (Figure 2). To exhibit the necessity of customizing HMM profiles from known UB/UBL regulators, here we compared the results in H. sapiens to the identifications in InterPro (24) and Pfam (23) databases (Supplementary Table S2). First, iUUCD 2.0 constructed HMM profiles for 58 families, whereas InterPro and Pfam only had HMM profiles for 48 and 43 families, respectively (Supplementary Table S2). Second, iUUCD 2.0 only obtained 1342 unique human regulators from the HMM-based identification, while InterPro and Pfam totally annotated 7599 and 2188 human proteins as UB/UBL regulators, separately (Supplementary Table S2). For the PHD family, there were 52 human E3 ubiquitin ligases annotated in iUUCD 2.0, whereas InterPro and Pfam identified 2744 (IPR013083) and 135 (PF00628) proteins containing at least one PHD domain, respectively (Supplementary Table S2). Then we used ‘CD-HIT’, a tool for clustering similar sequences (32), to evaluate the redundancy in InterPro and Pfam annotations. Using the clustering threshold of 100% identity, we found that 567 (20.7%) and 24 (17.8%) human PHD-containing proteins were redundantly annotated in InterPro and Pfam databases. Also, it was reported that PHD domains mainly function as either histone tail readers or E3 ligases (33,34). Thus, only a considerable proportion of PHD proteins would have E3 ligase activity. In addition, InterPro and Pfam annotations might miss a number of bona fide hits. For example, there were 55 human E3 ubiquitin ligases of the U-box family annotated in iUUCD 2.0, while InterPro and Pfam only contained 26 and 13 proteins, respectively (Supplementary Table S2). A well-characterized U-box protein, p33RUL/NOSIP (UniProt ID: Q9Y314) (35) was only correctly annotated in iUUCD 2.0. Taken together, our results demonstrated the procedure adopted in this study is important to ensure the data quality for the genome-wide identification.
For the families without HMM profiles, we conducted an orthologous search by using the reciprocal best hits (RBH) approach, which can efficiently identify orthologous pairs if two proteins in two different species reciprocally find each other as the best hit from (36). The blastall program in the BLAST package (37) was used for the sequence-based identification of orthologs. Combined with known and computationally identified UB/UBL regulators, we obtained 1230 E1s, 5636 E2s, 93 343 E3s, 9548 DUBs, 30 173 UBD proteins and 11 099 ULD proteins in 148 eukaryotes. A heatmap of member genes in all the families across eukaryotes was generated by the HemI program (http://hemi.biocuckoo.org/) (38) (Supplementary Figure S2), whereas the detailed data statistics are shown in Supplementary Table S3.
A multi-layer annotation of UB/UBL regulators
The iUUCD 2.0 database was developed as a gene-centred database. The classification and domain profile information were provided for each regulator, whereas a number of basic annotations such as Ensembl/UniProt accession numbers, gene/protein names, protein/nucleotide sequences, domain/motifs, functional descriptions and GO terms were taken from Ensembl (21) and UniProt (22) databases. For 58,889 UB/UBL enzymes in 148 eukaryotes, we also obtained the annotations of active sites for 7471 unique proteins (12.7%) from the UniProt database (22) (Supplementary Table S4). The primary references with PMIDs were present for known regulators, and computationally identified orthologs in other species were also integrated in the database if available.
To provide a more comprehensive resource, we first mapped all human regulators to 67 additional public databases such as The Cancer Genome Atlas (TCGA) (39), Catalog of Somatic Mutations in Cancer (COSMIC) (40), and International Cancer Genome Consortium (ICGC) (41) to obtain rich annotations that covered 11 aspects including cancer mutations, single nucleotide polymorphisms (SNPs), mRNA expression, DNA and RNA elements, protein–protein interactions (PPIs), protein 3D structures, disease-associated information, drug-target relations, PTMs, DNA methylation and protein expression/proteomics (Supplementary Table S5). In addition to H. sapiens, we further used the results from the 67 databases to annotate regulators in 7 other model organisms (Mus musculus, Rattus norvegicus, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Schizosaccharomyces pombe and Saccharomyces cerevisiae) (Supplementary Table S5). The PTM annotations in UniProt (22) were also included, whereas PTM sites annotated with ‘By similarity’, ‘Potential’ or ‘Probable’ were excluded for the integration. In total, we annotated 7138 UB/UBL regulators in eight species.
USAGE
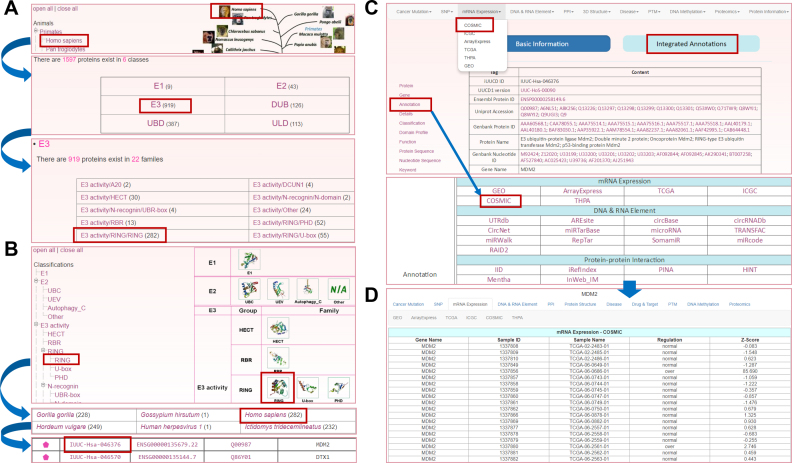
We developed the online service of iUUCD 2.0 to have an easy-to-use interface and implemented multiple options for querying the database. To depict the usage of iUUCD 2.0 here, we selected the human E3 ligase MDM2 as an example. Two options were provided to browse the database, either by species or by family classification. In the option of ‘Browse by species’, the left side represents the Ensembl taxonomic categories, whereas the right side represents the phylogenetic relationships of the eukaryotic species in Ensembl (21). Users can click ‘Homo sapiens’ and choose the ‘E3’ button, and the detailed family classification of human E3s will be presented (Figure 3A). Then, by pinpointing the human E3 activity/RING/RING family and clicking ‘IUUC-Hsa-046376’ (Figure 3A), the basic information page for human MDM2 can be viewed. Meanwhile, through the option of ‘Browse by classifications’, users can click the ‘RING’ on the left family tree or the structural picture of the RING family in the right-hand area. By choosing ‘Homo sapiens’ and ‘IUUC-Hsa-046376’ (Figure 3B), the users can also enter the basic information page for MDM2 (Figure 3C). In this page, the user will be able to view fundamental information including Ensembl Gene/Transcript/Protein ID, UniProt accession, family classification information, domain profiles, functional descriptions, and protein/nucleotide sequences (Figure 3C). To obtain more detailed annotations, users can either click on the navigation bar at ‘Integrated Annotations’ or the label of ‘Annotation’ (Figure 3C). The results will be shown in a new window. In addition, we implemented multiple search options including a simple search, batch search, advanced search, HMM search and BLAST search for using the database.
Figure 3.
Browsing the iUUCD 2.0. (A) The option to browse by species. (B) The option to browse by classification. The members of the human E3 activity/RING/RING family are shown in a tabular format. (C) The basic information on human MDM2 in the results page. (D) The multi-layer annotations of human MDM2.
DISCUSSION
Various UB/UBL conjugations are important protein PTMs and are involved in almost all aspects of biological processes and pathways (1–3). In addition to E1s, E2s, E3s and DUBs that modify or demodify protein substrates, a large number of proteins containing UBDs and/or ULDs also participate in the regulation of UB/UBL conjugation (7–14). In a previous study, we developed a family-based database, UUCD 1.0, that contained 56 949 E1s, E2s, E3s and DUBs in 70 eukaryotes with a data volume of ∼0.41 GB (15) (Supplementary Table S6). In this study, we further considered UBD and ULD proteins and integrated 136 512 UB/UBL regulators in 148 eukaryotes (Supplementary Table S6). We also provided rich annotations by mapping regulators in 8 model organisms to 67 additional public databases. The iUUCD 2.0 database contains a volume of ∼32.1 GB, which is a >75-fold increase. A detailed comparison of UUCD 1.0 and iUUCD 2.0 is shown in Supplementary Table S6.
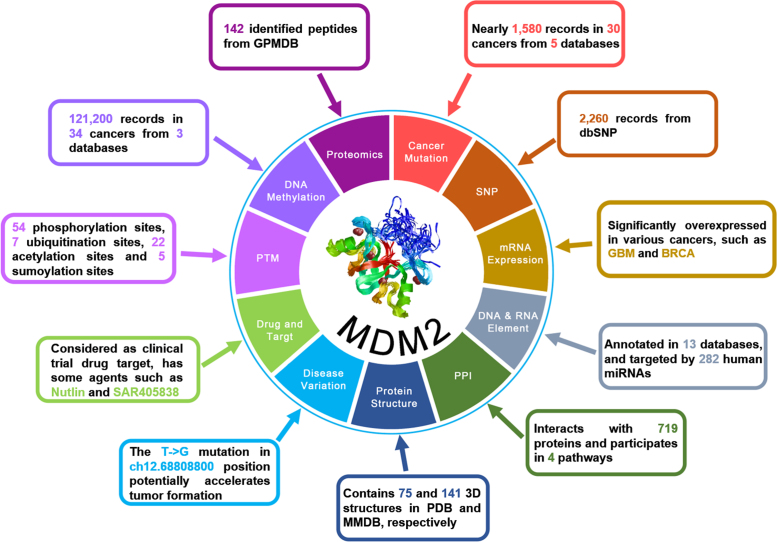
In recent years, advances in high-throughput techniques such as next-generation sequencing have generated an enormous amount of biological data. Together with extensive efforts from biocurators, many useful data resources have been constructed. The integration of multi-layer knowledge will undoubtedly provide rich annotations for a better understanding of UB/UBL conjugation. For example, 11 aspects of multi-dimensional annotations were summarized for human MDM2 (Figure 4). From the TCGA (39), COSMIC (40), ICGC (41) and analogous databases, we found nearly 1580 cancer mutation records from 30 cancers for MDM2, which is significantly overexpressed in a variety of cancers such as GBM and BRCA (42) (Figure 4). Human MDM2 also had 2260 SNP records from dbSNP (43) and could be targeted by 282 miRNAs (Figure 4). MDM2 interacts with 719 proteins and participates in 4 pathways, and there were 75 and 141 related 3D structures in PDB and MMDB, respectively (Figure 4). In particular, the T→G mutation in ch12.68808800 position annotated by ClinVar might accelerate tumor progression (44). Therefore, MDM2 has been widely accepted as a potential drug target for cancer therapy (45), and two such agents (Nutlin and SAR405838) annotated in DrugBank (46) and analogous databases are still in clinical trials. Moreover, MDM2 has 54 phosphorylation sites, 7 ubiquitination sites, 22 acetylation sites and 5 sumoylation sites, whereas 142 identified peptides were also obtained from GPMDB (47). In addition, the methylation information of MDM2 was also provided (Figure 4).
Figure 4.
The overview of multiple-layer annotations of human MDM2 integrated in iUUCD 2.0. A brief summary of the 67 additional public databases is given in Supplementary Table S5.
In the future, the iUUCD 2.0 database will be continuously updated and improved when new UB/UBL regulators are identified. Additionally, the classifications will be refined if new families are reported. More species will be added when their complete proteome and genome sets are available. In addition, we will include more annotations from other public databases to provide a more useful resource for UB/UBL conjugations.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Yaru Miao for her helpful comments during the database construction. American Journal Experts reviewed the manuscript prior to submission.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Special Project on Precision Medicine under the National Key R&D Program [2017YFC0906602]; Natural Science Foundation of China [31671360].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kerscher O., Felberbaum R., Hochstrasser M.. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006; 22:159–180. [DOI] [PubMed] [Google Scholar]
- 2. Swatek K.N., Komander D.. Ubiquitin modifications. Cell Res. 2016; 26:399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Komander D., Rape M.. The ubiquitin code. Annu. Rev. Biochem. 2012; 81:203–229. [DOI] [PubMed] [Google Scholar]
- 4. Popovic D., Vucic D., Dikic I.. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014; 20:1242–1253. [DOI] [PubMed] [Google Scholar]
- 5. Mendler L., Braun T., Muller S.. The ubiquitin-like SUMO system and heart function: from development to disease. Circ. Res. 2016; 118:132–144. [DOI] [PubMed] [Google Scholar]
- 6. Huang X., Dixit V.M.. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 2016; 26:484–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scheffner M., Nuber U., Huibregtse J.M.. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995; 373:81–83. [DOI] [PubMed] [Google Scholar]
- 8. Heride C., Urbe S., Clague M.J.. Ubiquitin code assembly and disassembly. Curr. Biol. 2014; 24:R215–R220. [DOI] [PubMed] [Google Scholar]
- 9. Buchberger A. From UBA to UBX: new words in the ubiquitin vocabulary. Trends Cell Biol. 2002; 12:216–221. [DOI] [PubMed] [Google Scholar]
- 10. Husnjak K., Dikic I.. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 2012; 81:291–322. [DOI] [PubMed] [Google Scholar]
- 11. Rajalingam K., Dikic I.. SnapShot: expanding the ubiquitin code. Cell. 2016; 164:1074–1074. [DOI] [PubMed] [Google Scholar]
- 12. Jentsch S., Pyrowolakis G.. Ubiquitin and its kin: how close are the family ties. Trends Cell Biol. 2000; 10:335–342. [DOI] [PubMed] [Google Scholar]
- 13. Schwartz D.C., Hochstrasser M.. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 2003; 28:321–328. [DOI] [PubMed] [Google Scholar]
- 14. Grabbe C., Dikic I.. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem. Rev. 2009; 109:1481–1494. [DOI] [PubMed] [Google Scholar]
- 15. Gao T., Liu Z., Wang Y., Cheng H., Yang Q., Guo A., Ren J., Xue Y.. UUCD: a family-based database of ubiquitin and ubiquitin-like conjugation. Nucleic Acids Res. 2013; 41:D445–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hutchins A.P., Liu S., Diez D., Miranda-Saavedra D.. The repertoires of ubiquitinating and deubiquitinating enzymes in eukaryotic genomes. Mol. Biol. Evol. 2013; 30:1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rose P.W., Prlic A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z. et al. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017; 45:D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrison J.S., Jacobs T.M., Houlihan K., Van Doorslaer K., Kuhlman B.. UbSRD: The Ubiquitin Structural Relational Database. J. Mol. Biol. 2016; 428:679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morreale F.E., Walden H.. Types of ubiquitin ligases. Cell. 2016; 165:248–248. [DOI] [PubMed] [Google Scholar]
- 20. Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R.. HMMER web server: 2015 update. Nucleic Acids Res. 2015; 43:W30–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al. Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The UniProt, C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.Y., Dosztanyi Z., El-Gebali S., Fraser M. et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017; 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coscoy L., Ganem D.. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 2003; 13:7–12. [DOI] [PubMed] [Google Scholar]
- 26. Taherbhoy A.M., Tait S.W., Kaiser S.E., Williams A.H., Deng A., Nourse A., Hammel M., Kurinov I., Rock C.O., Green D.R. et al. Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol. Cell. 2011; 44:451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma A., Malynn B.A.. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat. Rev. Immunol. 2012; 12:774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mergner J., Schwechheimer C.. The NEDD8 modification pathway in plants. Front. Plant Sci. 2014; 5:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abdul Rehman S.A., Kristariyanto Y.A., Choi S.Y., Nkosi P.J., Weidlich S., Labib K., Hofmann K., Kulathu Y.. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell. 2016; 63:146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hurley J.H., Lee S., Prag G.. Ubiquitin-binding domains. Biochem. J. 2006; 399:361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li W., Godzik A.. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006; 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- 33. Wang J., Muntean A.G., Wu L., Hess J.L.. A subset of mixed lineage leukemia proteins has plant homeodomain (PHD)-mediated E3 ligase activity. J. Biol. Chem. 2012; 287:43410–43416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matthews J.M., Bhati M., Lehtomaki E., Mansfield R.E., Cubeddu L., Mackay J.P.. It takes two to tango: the structure and function of LIM, RING, PHD and MYND domains. Curr. Pharm. Des. 2009; 15:3681–3696. [DOI] [PubMed] [Google Scholar]
- 35. Friedman A.D., Nimbalkar D., Quelle F.W.. Erythropoietin receptors associate with a ubiquitin ligase, p33RUL, and require its activity for erythropoietin-induced proliferation. J. Biol. Chem. 2003; 278:26851–26861. [DOI] [PubMed] [Google Scholar]
- 36. Tatusov R.L., Koonin E.V., Lipman D.J.. A genomic perspective on protein families. Science. 1997; 278:631–637. [DOI] [PubMed] [Google Scholar]
- 37. Coordinators N.R. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017; 45:D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deng W., Wang Y., Liu Z., Cheng H., Xue Y.. HemI: a toolkit for illustrating heatmaps. PLoS One. 2014; 9:e111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A. et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013; 502:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017; 45:D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang J., Baran J., Cros A., Guberman J.M., Haider S., Hsu J., Liang Y., Rivkin E., Wang J., Whitty B. et al. International Cancer Genome Consortium Data Portal–a one-stop shop for cancer genomics data. Database (Oxford). 2011; 2011:bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shaikh M.F., Morano W.F., Lee J., Gleeson E., Babcock B.D., Michl J., Sarafraz-Yazdi E., Pincus M.R., Bowne W.B.. Emerging role of MDM2 as target for anti-cancer therapy: a review. Ann. Clin. Lab. Sci. 2016; 46:627–634. [PubMed] [Google Scholar]
- 43. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol. Ther. 2003; 2:623–629. [PubMed] [Google Scholar]
- 46. Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014; 42:D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fenyo D., Beavis R.C.. The GPMDB REST interface. Bioinformatics. 2015; 31:2056–2058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.