Abstract
JASPAR (http://jaspar.genereg.net) is an open-access database of curated, non-redundant transcription factor (TF)-binding profiles stored as position frequency matrices (PFMs) and TF flexible models (TFFMs) for TFs across multiple species in six taxonomic groups. In the 2018 release of JASPAR, the CORE collection has been expanded with 322 new PFMs (60 for vertebrates and 262 for plants) and 33 PFMs were updated (24 for vertebrates, 8 for plants and 1 for insects). These new profiles represent a 30% expansion compared to the 2016 release. In addition, we have introduced 316 TFFMs (95 for vertebrates, 218 for plants and 3 for insects). This release incorporates clusters of similar PFMs in each taxon and each TF class per taxon. The JASPAR 2018 CORE vertebrate collection of PFMs was used to predict TF-binding sites in the human genome. The predictions are made available to the scientific community through a UCSC Genome Browser track data hub. Finally, this update comes with a new web framework with an interactive and responsive user-interface, along with new features. All the underlying data can be retrieved programmatically using a RESTful API and through the JASPAR 2018 R/Bioconductor package.
INTRODUCTION
Transcription factors (TFs) are sequence-specific DNA-binding proteins involved in the transcriptional regulation of gene expression (1). TFs bind to DNA through their DNA-binding domain(s) (DBDs), which are used for TF classification (2). DNA regions at which TFs bind are defined as TF-binding sites (TFBSs) and can be identified in vivo by methods such as chromatin immunoprecipitation (ChIP) or in vitro by methods based on binding of large pools of DNA fragments (e.g. Systematic evolution of ligands by exponential enrichment (SELEX) or protein-binding microarrays (PBM)) (reviewed in (3)). Analysis of TFBSs for a given TF provides models for its specific DNA-binding preferences, which in turn can be used to predict TFBSs in DNA sequences (4). This is important as experiments can only identify TFBSs that are bound in the cell and state analyzed.
The computational representation of TF binding preferences has evolved over the years, from simple consensus sequences to position frequency matrices (PFMs). A PFM summarizes experimentally determined DNA sequences bound by an individual TF by counting the number of occurrences of each nucleotide at each position within aligned TFBSs. Such matrices can be converted into position weight matrices (PWMs), also known as position-specific scoring matrices, which are probabilistic models that can be used to predict TFBSs in DNA sequences (reviewed in (5)).
PFMs/PWMs have been the standard models for describing binding preferences of TFs for many years. The JASPAR database is among the most popular and longest maintained databases for PFMs and a standard resource in the field. In particular, the JASPAR CORE collection of the database, which is the most used, stores non-redundant TF binding profiles, providing a single representative DNA binding model per TF decided by expert curators. Exceptionally, multiple TF-binding profiles are associated to a TF when it is known to interact with DNA with multiple distinct sequence preferences, due to differential splicing for example (6,7). JASPAR was created and persists under three guiding principles: (i) unrestricted open-access; (ii) manual curation and non-redundancy of profiles; and (iii) ease-of-use. The 2016 release of the JASPAR CORE collection stored 1082 non-redundant and manually curated TF-binding profiles as PFMs for TFs from six different taxonomic groups (vertebrates, plants, insects, nematodes, fungi and urochordata) (8).
An intrinsic limitation to PFMs/PWMs is that they ignore inter-nucleotide dependencies within TFBSs (9–13). TF–DNA interaction data derived from next-generation sequencing assays has improved the computational modeling of TF binding (14–19). For example, the TF flexible models (TFFMs) (14), based on first-order hidden Markov models, capture dinucleotide dependencies within TFBSs and were introduced in the 2016 release of the JASPAR database.
In this report, we describe the seventh release of JASPAR (8,20–24), which comes with a major expansion and update of the CORE collection of TF-binding profiles as PFMs and TFFMs. These models have been manually assessed by expert curators who reconciled recent high-throughput data with available literature and linked the models to the classification of their TF DBDs from TFClass (2). The CORE collection expansion is supported by a range of new functionalities and resources, including PFM clustering, genome-wide UCSC tracks of predicted TFBSs and fully redesigned user and programming interfaces.
EXPANSION AND UPDATE OF THE JASPAR CORE COLLECTION
In this 2018 release of the JASPAR database, we added 355 new PFMs for TFs from plants (270), vertebrates (84) and insects (1) to the JASPAR CORE collection (Table 1). Specifically, we added 322 PFMs (262 for plants, a 118% increase and 60 for vertebrates, an 11% increase) for TF monomers and dimers that were not previously present in JASPAR and updated 33 (8 in plants, 3% of JASPAR 2016, 24 in vertebrates, 5% of JASPAR 2016 and 1 in insects). The PFMs were manually curated using independent external literature supporting the candidate TF-binding preferences, as previously described in (23). The curated PFMs were derived from ChIP-seq (from ReMap (25) and (26–30)), DAP-seq (31), SMiLE-seq (32), PBM (33) and HT-SELEX (34) experiments. The JASPAR CORE collection now includes 1404 non-redundant PFMs (579 for vertebrates, 489 for plants, 176 for fungi, 133 for insects, 26 for nematodes and 1 for urochordata) (Table 1).
Table 1. Overview of the growth of the number of PFMs in the JASPAR 2018 CORE collection compared to the JASPAR 2016 CORE collection.
| Taxonomic group | Non-redundant PFMs in JASPAR 2016 | New non-redundant PFMs in JASPAR 2018 | Updated PFMs in JASPAR 2018 | Total PFMs (non-redundant) in JASPAR 2018 | Total PFMs (all versions) in JASPAR 2018 |
|---|---|---|---|---|---|
| Vertebrates | 519 | 60 | 24 | 579 | 719 |
| Plants | 227 | 262 | 8 | 489 | 501 |
| Insects | 133 | 0 | 1 | 133 | 140 |
| Nematodes | 26 | 0 | 0 | 26 | 26 |
| Fungi | 176 | 0 | 0 | 176 | 177 |
| Urochordata | 1 | 0 | 0 | 1 | 1 |
| Total | 1082 | 322 | 33 | 1404 | 1564 |
We continued with the incorporation of TFFM models, initiated in JASPAR 2016. In this release of JASPAR, we introduced 316 new TFFMs for vertebrates (95), plants (218) and Drosophila (3), which represents a 243% increase in the number of non-redundant TFFMs stored in the JASPAR CORE collection.
HIERARCHICAL CLUSTERING OF TF-BINDING PROFILES
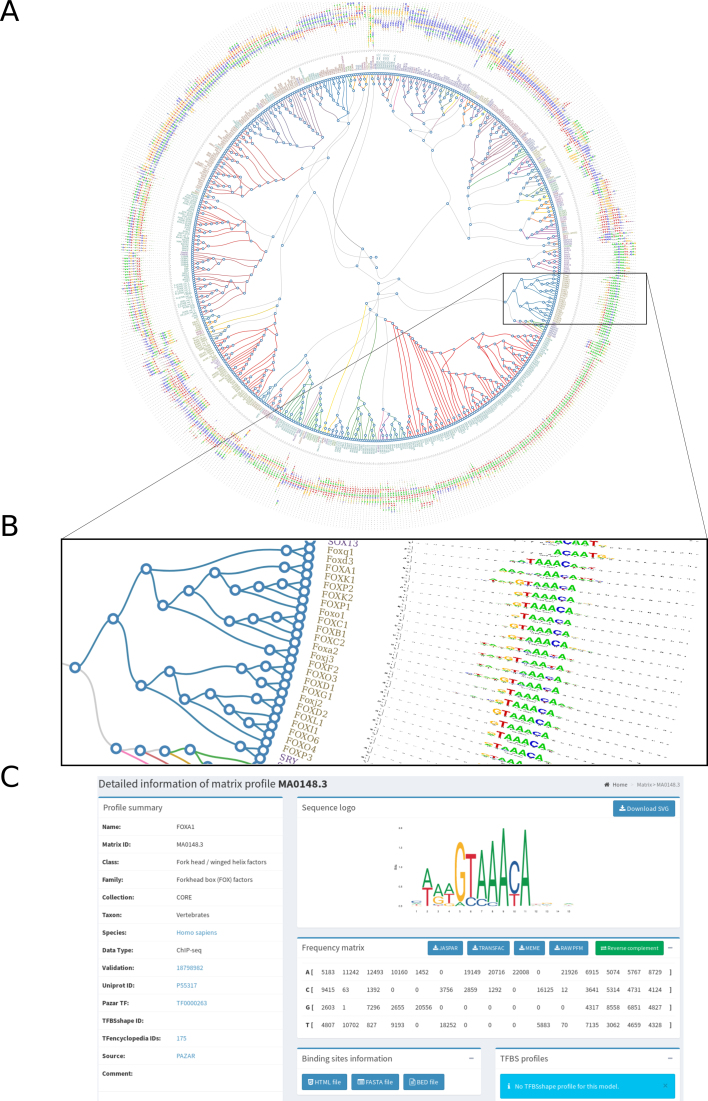
While the non-redundancy of binding profiles is one of the guiding principles of JASPAR, TFs with similar DBDs often have similar binding preferences (35,36). To facilitate the exploration of similar profiles in the JASPAR CORE collection, we performed hierarchical clustering of PFMs using the RSAT matrix-clustering tool (37). Specifically, the tool was applied to PFMs in each taxon independently as well as in each TF class per taxon. The clustering results are provided as radial trees (Figure 1), which can further be explored through dedicated web pages (http://jaspar.genereg.net/matrix-clusters).
Figure 1.
JASPAR PFM clustering. (A) Radial tree representing the clusterization of the JASPAR CORE vertebrate PFMs. (B) Zoom in view of the radial tree where the predicted clusters are highlighted at the branches and the TF classes are indicated with different colors at the leaves. (C) Clicking on a leaf in the radial tree will open a link to the corresponding motif description page on the JASPAR website (the MA0148.3 profile associated to FOXA1 is provided here as an example).
JASPAR UCSC TRACKS FOR GENOME-WIDE ANALYSES OF TFBSs
A typical application of JASPAR TF-binding profiles in gene regulation studies is the identification of TFBSs in DNA sequences for further analyses. Although, we recognize that genome-wide PWM-based predictions contain a high number false positives, we believe that they are a powerful resource for the research community in the context of a variety of genomic information, including transcription start site activity, DNA accessibility, histone marks, evolutionary conservation or in vivo TF binding (38–46). To facilitate such integrative analyses, we have performed TFBS predictions on the human genome using the JASPAR CORE vertebrate PFMs (see Supplementary Data for details on the computation). The predicted TFBSs are publicly available through a UCSC Genome Browser data hub (47) containing tracks for the human genome assemblies hg19 and hg38 (http://jaspar.genereg.net/genome-tracks/).
A NEW, POWERFUL AND USER-FRIENDLY WEB INTERFACE
A new web interface
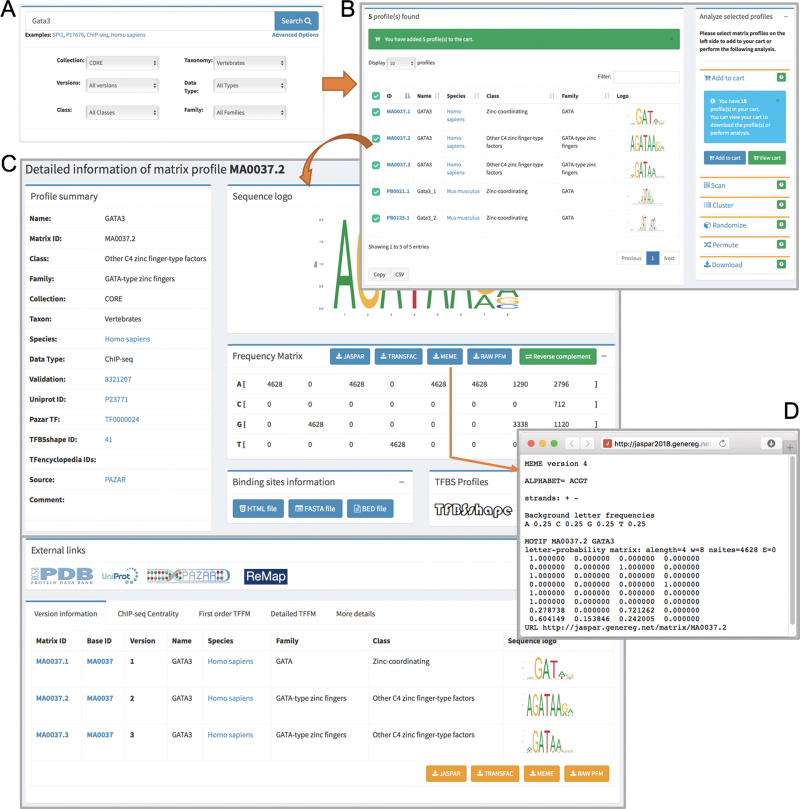
The JASPAR 2018 release comes with a completely redesigned web interface that meets modern web standards. This interactive web framework is implemented using Django, a model-view-controller based web-framework for Python. We used MySQL as a backend database to store profile metadata and Bootstrap as a frontend template engine. We have greatly improved the visibility and usability of existing functionality, created easier navigation with semantic URLs, and enhanced browsing and searching. On the homepage, we provide a dynamic tour of JASPAR 2018, walking users through the main features of the new website. A video of the tour is available at http://jaspar.genereg.net/tour. The database can be browsed for individual collections by using the navigation links on the left sidebar. Moreover, it can be searched for each of the six different taxonomic groups included in the JASPAR CORE collection using the tabs available on the homepage (Figure 2). TF-binding profiles can be further filtered through the case insensitive search option available on the homepage. In addition, through the ‘Advanced Options’, the search criteria can be further restricted (Figure 2A). Search results are presented in a responsive and paginated table along with sequence logos of the PFMs, which can be selected for download or to perform a variety of analyses available on the right panel (Figure 2B). All information in the tables can be downloaded as comma-separated value files. Profile IDs and sequence logos can be clicked to view the detailed profile pages (Figure 2C). PFMs can be downloaded in several formats including JASPAR, TRANSFAC and MEME (Figure 2D). Furthermore, we have incorporated new features to the web interface, such as ‘Add to Cart’, where users can add TF profiles of interest for download or further analyses (Figure 2B). Finally, we have introduced semantic URLs to facilitate external linking to the detailed pages of individual profiles (e.g. http://jaspar.genereg.net/matrix/MA0059.1/). We have implemented a URL redirection mechanism to correctly direct the links pointing to previous JASPAR URL patterns from external resources.
Figure 2.
Overview of the JASPAR 2018 new web interface with interactive searching activity. (A) A quick and detailed search feature on the homepage. (B) A responsive table lists the searched profile(s), which can be further selected and added to the cart listed on the right panel for users to perform their own analyses. (C) A detailed page for the GATA3 matrix profile, which is divided into sub-panels including the profile summary, sequence logo, PFM, TF-binding information, external links, version information, ChIP-seq centrality, TFFM and other details. (D) The PFM for the GATA3 profile (MA0037.2) is downloaded in MEME format using the RESTful API.
RESTful API
In previous releases, the underlying data could be retrieved as flat files or by using programming language-specific modules. Associated with this release, we introduced a RESTful API to access the JASPAR database programmatically (see https://www.biorxiv.org/content/early/2017/07/06/160184 for details). The RESTful API enables programmatic access to JASPAR by most programming languages and returns data in seven widely used formats: JSON, JSONP, JASPAR, MEME, PFM, TRANSFAC and YAML. Further, it provides a browsable interface and access to the JASPAR motif inference tool for bioinformatics tool developers. The RESTful API is implemented in Python using the Django REST Framework and is freely accessible at http://jaspar.genereg.net/api/. The source code for the website and RESTful API are freely available at https://bitbucket.org/CBGR/jaspar under GPL v3 license.
CONCLUSION AND PERSPECTIVES
In this seventh release of the JASPAR database, we continue our commitment to provide the research community with high-quality, non-redundant TF-binding profiles for TFs in six taxa. As in previous releases, we have greatly expanded the number of available profiles in the database, both for PFMs and TFFMs. We also greatly improved user experience through a new easy-to-use website and a RESTful API that grants universal programmatic access to the database. Moreover, for the PFMs in the JASPAR CORE collection, we provide a hierarchical clustering and genome-wide TFBS predictions for the hg19 and hg38 human genome assemblies as UCSC tracks.
During the curation process, hundreds of PFMs were discarded because our curators failed to find any support from existing literature. As new experiments and data become available, binding preferences for these TFs will be considered for JASPAR incorporation. For instance, we re-examined data from (34) to incorporate seven previously excluded PFMs into JASPAR 2018. In the future, we would like to engage the scientific community in the curation process to increase our capacity to introduce new TF-binding profiles in JASPAR. We plan to dedicate a specific section of the website to hosting the profiles that were not introduced into JASPAR, to encourage researchers to perform experiments and/or point us to literature that our curators missed in order to support these profiles. We believe that the engagement of the scientific community to support JASPAR will further improve our capacity to expand the collection of high quality TF-binding profiles.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the scientific community for performing experimental assays of TF–DNA interactions and for publicly releasing the data. We thank Georgios Magklaras and his team for IT support. We thank José Manuel Franco for sharing the plant PBM data, Jens De Ceukeleire for help with plant ChIP-seq data processing and José Luis Villanueva-Cañas for sharing the Drosophila TFFMs prior to publication. We thank Rachelle Farkas for proofreading the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Norwegian Research Council, Helse Sør-Øst, and University of Oslo through the Centre for Molecular Medicine Norway (NCMM) (to A.M., M.G., A.K.); Genome Canada and Canadian Institutes of Health Research (OnTarget Grants) [255ONT and BOP-149430 to W.W.W., O.F., R.v.d.L., D.J.A.]; Natural Sciences and Engineering Research Council of Canada (Discovery Grant) [RGPIN-2017–06824 to W.W.W.]; Weston Brain Institute [20R74681 to O.F.]; Agence Nationale de la Recherche [ANR-10-LABX-49–01 to F.P., A.S.]; IDEX graduate schoool (to A.S.); CNRS (to A.B., F.P.); Research Foundation–Flanders Grant [G001015N to S.R.K.]; French Ministry of Higher Education and Research (MESR) PhD Fellowship (to J.A.C.-M.); Lundbeck Foundation (to A.S.); Independent Research Fund Denmark (to A.S.); Innovation Fund Denmark (to A.S.); Elixir Denmark (to A.S.); Wellcome Trust [106954 to G.T., D.B., B.L.]; Biotechnology and Biological Sciences Research Council [BB/N023358/1 to G.T., D.B., B.L.]; Medical Research Council UK [MC_UP_1102/1 to G.T., D.B., B.L.]. The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M.. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009; 10:252–263. [DOI] [PubMed] [Google Scholar]
- 2. Wingender E., Schoeps T., Haubrock M., Dönitz J.. TFClass: a classification of human transcription factors and their rodent orthologs. Nucleic Acids Res. 2015; 43:D97–D102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xie Z., Hu S., Qian J., Blackshaw S., Zhu H.. Systematic characterization of protein-DNA interactions. Cell. Mol. Life Sci. 2011; 68:1657–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wasserman W.W., Sandelin A.. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004; 5:276–287. [DOI] [PubMed] [Google Scholar]
- 5. Stormo G.D. Modeling the specificity of protein-DNA interactions. Quant. Biol. 2013; 1:115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stormo G.D. DNA motif databases and their uses. Curr. Protoc. Bioinformatics. 2015; 51:1–6. [DOI] [PubMed] [Google Scholar]
- 7. Badis G., Berger M.F., Philippakis A.A., Talukder S., Gehrke A.R., Jaeger S.A., Chan E.T., Metzler G., Vedenko A., Chen X. et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009; 324:1720–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mathelier A., Fornes O., Arenillas D.J., Chen C.-Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R. et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016; 44:D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Man T.K., Stormo G.D.. Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res. 2001; 29:2471–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bulyk M.L., Johnson P.L.F., Church G.M.. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 2002; 30:1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou Q., Liu J.S.. Modeling within-motif dependence for transcription factor binding site predictions. Bioinformatics. 2004; 20:909–916. [DOI] [PubMed] [Google Scholar]
- 12. Tomovic A., Oakeley E.J.. Position dependencies in transcription factor binding sites. Bioinformatics. 2007; 23:933–941. [DOI] [PubMed] [Google Scholar]
- 13. Chin F., Leung H.C.M.. DNA motif representation with nucleotide dependency. IEEE/ACM Trans. Comput. Biol. Bioinform. 2008; 5:110–119. [DOI] [PubMed] [Google Scholar]
- 14. Mathelier A., Wasserman W.W.. The next generation of transcription factor binding site prediction. PLoS Comput. Biol. 2013; 9:e1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zellers R.G., Drewell R.A., Dresch J.M.. MARZ: an algorithm to combinatorially analyze gapped n-mer models of transcription factor binding. BMC Bioinformatics. 2015; 16:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eggeling R., Roos T., Myllymäki P., Grosse I.. Inferring intra-motif dependencies of DNA binding sites from ChIP-seq data. BMC Bioinformatics. 2015; 16:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Siebert M., Söding J.. Bayesian Markov models consistently outperform PWMs at predicting motifs in nucleotide sequences. Nucleic Acids Res. 2016; 44:6055–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mathelier A., Xin B., Chiu T.-P., Yang L., Rohs R., Wasserman W.W.. DNA shape features improve transcription factor binding site predictions in vivo. Cell Syst. 2016; 3:278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Omidi S., Zavolan M., Pachkov M., Breda J., Berger S., van Nimwegen E.. Automated incorporation of pairwise dependency in transcription factor binding site prediction using dinucleotide weight tensors. PLoS Comput. Biol. 2017; 13:e1005176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sandelin A., Alkema W., Engström P., Wasserman W.W., Lenhard B.. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004; 32:D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vlieghe D., Sandelin A., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B.. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006; 34:D95–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bryne J.C., Valen E., Tang M.-H.E., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A.. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008; 36:D102–D106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Portales-Casamar E., Thongjuea S., Kwon A.T., Arenillas D., Zhao X., Valen E., Yusuf D., Lenhard B., Wasserman W.W., Sandelin A.. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010; 38:D105–D1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.-Y., Chou A., Ienasescu H. et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014; 42:D142–D1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chèneby J., Gheorghe M., Artufel M., Mathelier A., Ballester B.. ReMap 2018: An updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seqexperiments. Nucleic Acids Res. 2017; doi:10.1093/nar/gkx1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eveland A.L., Goldshmidt A., Pautler M., Morohashi K., Liseron-Monfils C., Lewis M.W., Kumari S., Hiraga S., Yang F., Unger-Wallace E. et al. Regulatory modules controlling maize inflorescence architecture. Genome Res. 2014; 24:431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verkest A., Abeel T., Heyndrickx K.S., Van Leene J., Lanz C., Van De Slijke E., De Winne N., Eeckhout D., Persiau G., Van Breusegem F. et al. A generic tool for transcription factor target gene discovery in Arabidopsis cell suspension cultures based on tandem chromatin affinity purification. Plant Physiol. 2014; 164:1122–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li C., Qiao Z., Qi W., Wang Q., Yuan Y., Yang X., Tang Y., Mei B., Lv Y., Zhao H. et al. Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of opaque2 in maize. Plant Cell. 2015; 27:532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cui X., Lu F., Qiu Q., Zhou B., Gu L., Zhang S., Kang Y., Cui X., Ma X., Yao Q. et al. REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 2016; 48:694–699. [DOI] [PubMed] [Google Scholar]
- 30. Birkenbihl R.P., Kracher B., Somssich I.E.. Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell. 2017; 29:20–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O’Malley R.C., Huang S.-S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R.. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016; 165:1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Isakova A., Groux R., Imbeault M., Rainer P., Alpern D., Dainese R., Ambrosini G., Trono D., Bucher P., Deplancke B.. SMiLE-seq identifies binding motifs of single and dimeric transcription factors. Nat. Methods. 2017; 14:316–322. [DOI] [PubMed] [Google Scholar]
- 33. Franco-Zorrilla J.M., López-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R.. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G. et al. DNA-binding specificities of human transcription factors. Cell. 2013; 152:327–339. [DOI] [PubMed] [Google Scholar]
- 35. Weirauch M.T., Yang A., Albu M., Cote A., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014; 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sandelin A., Wasserman W.W.. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004; 338:207–215. [DOI] [PubMed] [Google Scholar]
- 37. Castro-Mondragon J.A., Jaeger S., Thieffry D., Thomas-Chollier M., van Helden J.. RSAT matrix-clustering: dynamic exploration and redundancy reduction of transcription factor binding motif collections. Nucleic Acids Res. 2017; 45:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kwon A.T., Arenillas D.J., Worsley Hunt R., Wasserman W.W.. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3. 2012; 2:987–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mathelier A., Lefebvre C., Zhang A.W., Arenillas D.J., Ding J., Wasserman W.W., Shah S.P.. Cis-regulatory somatic mutations and gene-expression alteration in B-cell lymphomas. Genome Biol. 2015; 16:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verfaillie A., Imrichova H., Janky R., Aerts S.. iRegulon and i-cistarget: reconstructing regulatory networks using motif and track enrichment. Curr. Protoc. Bioinformatics. 2015; 52:1–39. [DOI] [PubMed] [Google Scholar]
- 41. Arenillas D.J., Forrest A.R.R., Kawaji H., Lassmann T. FANTOM Consortium . FANTOM Consortium Wasserman W.W., Mathelier A.. CAGEd-oPOSSUM: motif enrichment analysis from CAGE-derived TSSs. Bioinformatics. 2016; 32:2858–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi W., Fornes O., Mathelier A., Wasserman W.W.. Evaluating the impact of single nucleotide variants on transcription factor binding. Nucleic Acids Res. 2016; 44:10106–10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drabløs F., Lennartsson A., Rönnerblad M., Hrydziuszko O., Vitezic M. et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015; 347:1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. FANTOM Consortium and the RIKEN PMI and CLST (DGT) Forrest A.R.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J.L., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M. et al. A promoter-level mammalian expression atlas. Nature. 2014; 507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Neph S., Vierstra J., Stergachis A.B., Reynolds A.P., Haugen E., Vernot B., Thurman R.E., John S., Sandstrom R., Johnson A.K. et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012; 489:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. et al. The accessible chromatin landscape of the human genome. Nature. 2012; 489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T., Nguyen N., Paten B., Zweig A.S., Karolchik D. et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014; 30:1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.