Abstract
Web repositories for almost all ‘omics’ types have been generated—detailing the repertoire of representatives across different tissues or cell types. A logical next step is the combination of these valuable sources. With IMOTA (interactive multi omics tissue atlas), we developed a database that includes 23 725 relations between miRNAs and 23 tissues, 310 932 relations between mRNAs and the same tissues as well as 63 043 relations between proteins and the 23 tissues in Homo sapiens. IMOTA also contains data on tissue-specific interactions, e.g. information on 331 413 miRNAs and target gene pairs that are jointly expressed in the considered tissues. By using intuitive filter and visualization techniques, it is with minimal effort possible to answer various questions. These include rather general questions but also requests specific for genes, miRNAs or proteins. An example for a general task could be ‘identify all miRNAs, genes and proteins in the lung that are highly expressed and where experimental evidence proves that the miRNAs target the genes’. An example for a specific request for a gene and a miRNA could for example be ‘In which tissues is miR-34c and its target gene BCL2 expressed?’. The IMOTA repository is freely available online at https://ccb-web.cs.uni-saarland.de/imota/.
INTRODUCTION
High-throughput approaches lead to the development of multiple resources in which different ‘omics’ types information on tissue specificity of respective representatives of transcriptomics, proteomics or epigenomic data are available. A current trend is to create databases that integrate information from multiple resources to provide users with an as complete as possible picture on molecular processes, which are partially specific for certain tissue types.
In the following, we mention and briefly describe selected databases that have been built from high-throughput omics datasets and that host information on the tissue specificity of comics datasets. This list is not thought to be a complete enumeration and review of all available respective resources but rather to show frequently used solutions as examples.
The ‘Expression Atlas’ hosted by the EMBL-EBI (The European Molecular Biology Laboratory - European Bioinformatics Institute) (1) (http://www.ebi.ac.uk/gxa/) is an open resource that allows users to find information about gene and protein expression across many different species and diverse biological conditions (e.g. different tissues, cell types, developmental stages or even diseases). The intuitive handling and concise representation of the EBI expression atlas were partially used as the prototype for developing IMOTA (interactive multi omics tissue atlas). A large fraction of data contained in this resource are derived from the ArrayExpress dataset, which is also maintained by the EMBL. It also includes data from the human protein atlas (2,3) (version 15), ‘The Genotype-Tissue Expression’ (4) (GTEx) project and others.
‘ProteomicsDB’ (5) is a database which allows browsing the human proteome, including protein expressions and peptide identification values. It was jointly developed by the Technische Universität München, SAP (Systems, Applications & Products in Data Processing) and GSK (GlaxoSmithKline). ProtenomicDB is based on the SAP HANA platform for data mining and visualization. It contains information from more than 400 experiments and roughly 80 projects, covering 80% of the human proteome.
The ‘Human Epigenome Atlas’ includes human reference epigenomes and the results of their integrative and comparative analysis. It has been developed by the Baylor College.
The ‘Human miRNA tissue atlas’ (6) describes the expression rates of miRNAs in over 30 solid tissues. A special feature of this resource is that the miRNA patterns have been measured from the same corpses in order to minimize the effect of differences between different individuals. It is also possible to compare the expression of a specific miRNA with data from other experiments such as the Gene Expression Omnibus (7) series, which contains expression profiles for different human tissues, disease conditions or body fluids. Also data on ancient miRNA expression from the Tyrolean Iceman are included. The human miRNA tissue atlas was developed at Saarland University.
The web tool ‘SlideBase’ (8) allows the user to select a subset of genes, miRNAs and proteins by defining custom expression level thresholds for given cell types and tissues with sliders. It includes data from BioGPS (9) (http://biogps.org/), functional annotation of mammals (10) (http://fantom.gsc.riken.jp/), GTEx (http://www.gtexportal.org/) and aforementioned ‘human protein atlas’ (http://www.proteinatlas.org/). These databases can be filtered individually to select data and was implemented by the University of Copenhagen in cooperation with RIKEN Center for Life Science Technologies.
Although different of the aforementioned resources have already the ambition to describe data of more than only one omics type, there is a clear trend and need for a sophisticated solution that integrates tissue expression of relevant omics types and also the tissue specificity of the interaction between them. With IMOTA, we developed a database that integrates expression profiles of 1353 miRNAs, 18 206 genes and 4422 proteins in 23 solid tissues. IMOTA also contains data on tissue-specific interactions, e.g. information on 331 413 miRNAs and target gene pairs that are jointly expressed in the 23 tissues. Further, it includes 23 725 relations between miRNAs and the 23 tissues, 310 932 relations between mRNAs and the same tissues as well as 63 043 relations between proteins and the 23 tissues. The content and functionality of IMOTA is described in details in the following section. Thereafter, the database set-up and information on the implementation is provided. By using the intuitive filter techniques, it is—with minimal effort—possible to answer various questions. Selected examples are given in the ‘Database Functionality and Example Applications’ section. Finally, we describe how we evaluated the usability of the database, name limitations and future directions of IMOTA.
An important aspect of our effort to integrate more specific resources is that we do not aim to make the respective detailed databases obsolete. IMOTA rather is thought to be a high level entry point to the specific repositories. Thus, we pay attention not only to acknowledge the work of others but also to directly link to the original sources wherever possible.
DATABASE CONTENT
IMOTA relies on the work of different research groups. Generally, the data resources can be divided in two parts. First, background databases that contain general information on genes, proteins, miRNAs, targets of miRNAs or tissues. Second, databases that store the actual expression profiles of the omics data in tissues.
Among the background databases, we include data from NCBI gene repository (https://www.ncbi.nlm.nih.gov/gene) to get the relevant information on genes and proteins. Information on organs is included from the Ontology Lookup Service of EMBL-EBI (https://www.ebi.ac.uk/ols/). For miRNAs, two different databases are included, the current gold standard repository miRBase (11) (http://mirbase.org) and miRCarta (http://www.ccb.uni-saarland.de/mircarta/), an up-to-date resource storing information on miRBase miRNAs as well as candidates that have been discovered from various Next-Generation Sequencing (NGS) experiments (manuscript in preparation). Information on targets of miRNAs are integrated from the miRTarBase (12) (http://mirtarbase.mbc.nctu.edu.tw) and targetscan (13) (http://www.targetscan.org/vert_71/).
To include tissue based expression profiles, the human protein atlas (version 16, http://www.proteinatlas.org) that includes protein expression and localization profiles for a large majority of all protein-coding genes based on both RNA and protein data in normal tissues, cancer and cell lines that occur in the human body was used. This expression atlas is divided into three parts: cancer, cell and normal tissue; including more than 10 million corresponding images. The normal tissue atlas contains data and images for protein and mRNA expression and distribution across organs and tissues in the human body. The (quantitative) protein data covers distribution and expression rates on tissue and cell level (about 76 different cell types), while the mRNA data focuses on tissues. The miRNA data were included from the human miRNA tissue atlas (https://ccb-web.cs.uni-saarland.de/tissueatlas/) that provides data on 1997 miRNAs in 31 solid human tissues. The data were extracted from two male individuals (postmortem) to minimize the risk of inter-individual variability.
DATABASE SET-UP, IMPLEMENTATION AND VISUALIZATION
The database itself is a MySQL database. The entity relationship diagram of the IMOTA database design is shown in Supplemental Figure S1. The web user interface uses HTML, written with the templating language pug, for displaying pages, CSS/Bootstrap for styling and JavaScript along with several libraries to support interactive page components such as charts and the anatomy model. The server-side application logic was implemented with Django and Python. To provide interactivity, IMOTA utilizes a set of JavaScript libraries that use Ajax calls to communicate with the back end. By using asynchronous calls in the background, the experience is not interrupted by reloading. The interconnected graphs are implemented with the JavaScript library dc.js. The library uses crossfilter.js as calculating engine to explore large multivariate datasets in web browsers and d3.js to render charts. The overall system architecture is shown schematically in Supplemental Figure S2. The auto-complete capabilities of the search boxes were implemented with twitter’s typeahead engine, a fast and robust suggestion engine. All the visualization in IMOTA are Scalable Vector Graphics (SVG). As SVG is a markup language like HTML, each component of the SVG is represented as a separate element in the document and can be individually addressed. The SVG format allows to interact with graphics in the same way as any other HTML element.
For visualization, two main widgets were implemented. First, an interface to browse, query and download the data to get a general overview, called the interactive overview (IO view). Second, a site to query the data for miRNA–target interaction relationships (MTI view). Both are described in the next section.
IMOTA also includes a visualization of the human body. Here, the male and female version of the expression atlas anatomogram were combined and organs not included in our atlas were removed. In the MTI view, the colors of the tissues in the visualization correspond to the colors of the heat map cells and in the IO view they reflect colors of the ‘Count per Tissue’ chart.
A very important aspect of IMOTA was to be self-explanatory and applicable even for non-experts with minimal effort. This is enabled not only by a tutorial and help page but also by tool tips that can be used at any step to get explanations on the current functionality and respective results.
DATABASE FUNCTIONALITY AND EXAMPLE APPLICATIONS
Interactive overview (IO)
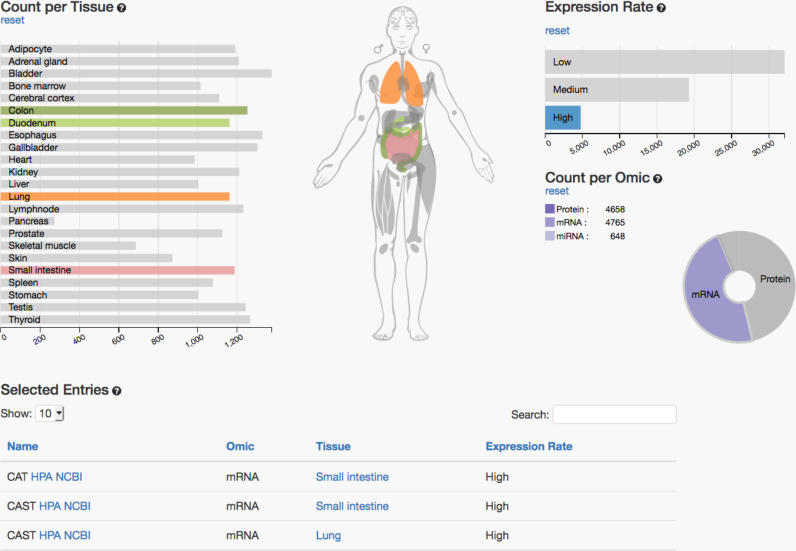
On the IO page, data can be filtered through interaction with the charts and the search fields. Specifically, the IO view contains three diagrams that are directly linked to each other and to the anatomic model in the middle (see Figure 1). Each of the three interactive diagrams generally represents one filtering option.
The ‘Expression Rate’ bar chart displays how many entries in the database are expressed at a certain level. For this task, the expression intensities have been discretized to ‘expression rates’ in the following categories: not expressed, low, medium or high expressed. The protein and gene expression values were integrated from the human protein atlas. For the RNASeq data, the number of fragments per kilobase gene model and million reads (FPKM) was used and the thresholds were as followings: 0–0.5 FPKM was considered not detected, 0.5–10 FPKM as low, 10–50 as medium and above 50 FPKM as high expressed. For the miRNA data—which have been measured by microarray technology—we calculated the following thresholds by a histogram-based approach: intensities below 1 were considered not detected, from 1–10 as low, from 10–500 as medium and above 500 counts as high expressed.
The ‘Count per Omic’ pie chart visualizes the proportional count of the individual omics data, i.e. how many genes, proteins and miRNAs are considered in the current selection in each of the organs.
The ‘Count per Tissue’ bar chart resolves omic counts per tissue according to the user’s selection. By clicking on a part of the chart, the underlying data are filtered and the results are displayed directly within the three charts, as well as in the table at the bottom of the page. The currently filtered information that is visualized on top and detailed in the table at the bottom of the page can, at each point, be downloaded in CSV format.
Figure 1.
Interactive overview of interactive multi omics tissue atlas. The left panel shows the organs as bar chars. Selected organs can be en/disabled by clicking either on the anatomic model or the bars. The right panel shows available omics types. By clicking the pie chart different omics datasets can be selected or deselected. The top right part shows the expression level. As for the other parameters, different levels can be selected. At the bottom the list of currently matching items is presented. This list can be downloaded as CSV file.
As an example we present in Figure 1 the analysis how many and which genes and miRNAs are highly expressed in lung, colon, small intestine and duodenum. Each gray-colored part in the interactive visualization can be clicked to include or exclude the respective condition. Immediately the information is updated and the newly selected results are presented. In the IO view, the visualization of the human body is interconnected with the related tissue diagram and will also be updated automatically if the user includes or excludes certain tissues, omics data or expression rates.
Searching for a gene or a miRNA with the search fields, filters the data depending on the search input and displays the information in the charts. For more detailed information about the relationships between omics profiles in the different tissues, the MTI view is however more appropriate. With the IO view the user can gain a quick overview over the data included in IMOTA.
To use discrete expression rates as described above has certain advantages. They allow for an easy filtering and concise representation of the results. Further, the categorization is done independent of the tissue, i.e. the thresholds have not been adjusted according to different tissues. While this consideration is well suited for getting a first overview and for filtering purposes, detailed analysis requires a closer look at the data. To this end, we also provide links to the original data resources in the results table below the diagrams (see Figure 1). This facilitates to compare the original expression values to each other. As example, a miRNA that is considered highly expressed in lung such as let-7a-5p can still be expressed orders of magnitudes higher in other tissues. Here, also the influence of different normalization techniques has to be taken into account. For our own miRNA tissue atlas, we thus present in addition to the raw data values also quantile normalized expression intensities as well as intensities following variance stabilization that has shown high performance on microarray data (14).
MTI view
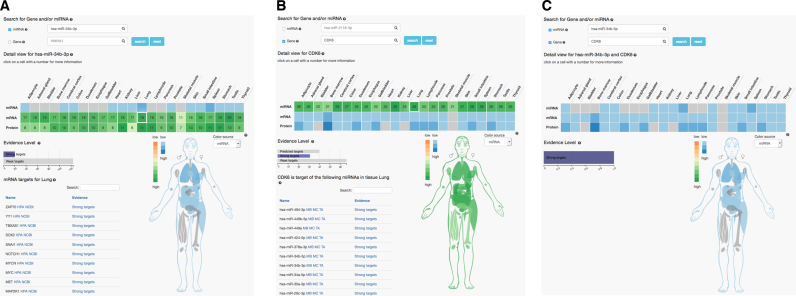
The miRNA–target interaction view displays further information about target relations and evidence levels between miRNAs, mRNAs and proteins. A heat map represents the relationships between the three omics datasets, their expression level per tissue and the number of miRNA targets or gene sources per tissue. For a search by miRNA, the heat map displays the miRNA expression rates per tissue in the first row and the number of target genes per tissue divided by omic type (mRNA, protein) in the other two rows. Furthermore, the evidence level bar chart displays the overall distribution of evidence levels for the displayed targets. To filter the heat map by evidence level, the respective bar (predicted targets, strong targets or weak targets) can be clicked. A table shows the selected miRNA targets or gene sources for a specific cell of the heat map. In the MTI view, the body model is connected to the heat map. The color source can be freely changed from miRNA to mRNA or protein.
As an example the results for miR-34b-3p in the MTI view are presented in Figure 2A. The miRNA is expressed in 14 different tissues (blue highlighted cells). The same colors are used to highlight the organs in the anatomic model. For all tissues, the target genes and corresponding proteins of this miRNA are shown in different shades of green. By selecting gene or protein from the drop down, the organs can be colored accordingly. In the current representation, the entry lung/gene is selected and highlighted. This means that in the current results table all genes that are targets of miR-34b-3p and are expressed in the lung are enumerated and links to the human protein atlas with detailed expression intensity values as well as links to the NCBI are provided.
Figure 2.
Three miRNA–target interaction relationships views. The left part shows the miRNA centric results, the middle part gene centric results and the right part results specific for one pair of a miRNA/gene.
Analogously, the results for the gene CDK6 can be displayed (Figure 2B). In the representation, the miRNAs in the lung are selected and accordingly displayed in the table below the graphic. This table also contains the miRNA from the previous example, miR-34b-3p.
In the last example, the most specific type of question is addressed. In which tissues are miR-34b-3p and its target gene CDK6 jointly expressed. In 12 tissues, miRNA, gene and the corresponding protein are found (Figure 2C). Only in the skeletal muscle neither the gene, nor the protein or miRNA are detected.
Although, we observed a significant enrichment of ‘true’ target genes of miRNAs that have been validated by reporter assays with increasing number of tissues where miRNA, gene and protein are jointly expressed, a direct comparison of the regulatory patterns between different organs is challenging. It may be that a gene is higher expressed in one organ compared to a second organ although a miRNA regulating this gene is also higher in the first organ simply because in the second organ other miRNAs targeting the gene are higher expressed as compared to the first organ. Other reasons for the presence of protein products even in case of a successful repression of a gene exist, e.g. if the miRNA targets only one splice form of the gene.
Nonetheless, the application scenarios highlight how many different tasks can be fulfilled by using IMOTA providing useful insights in the co-expression of miRNAs, genes and proteins. From very general things, such as, the enumeration of all miRNAs expressed in one or several tissues up to the very detailed information whether a certain miRNA regulates a specific gene in one tissue.
Link to detailed sources, updates
IMOTA relies on many different data resources that have been described in the database content section. Our ambition is to provide life scientists an easy entry to the complex and specific datasets, especially if per-filtering and combined consideration of different omics datasets is required. Thus, we pay attention to provide links to the original data sources wherever possible. For miRNAs, three links to other external resources are provided: to the miRBase, miRCarta and the miRNA tissue atlas. Likewise, genes are linked with the tissue atlas and NCBI gene. Tissues are linked to the EMBL-EBI Ontology Lookup Service. miRNA/target gene interactions are connected to the original entry from miRTarbase. If for example detailed expression levels or information on which experimental technique was used to identify the target are requested, IMOTA users can identify them without additional work by going to the original repositories. Besides ensuring that the resources get the deserved credits, this way of presenting the data has also the advantage to facilitate easy database update. Whenever one resource is updated the information is extracted and all connections to the respective resources are semi-automatically build.
TESTING, LIMITATIONS, FUTURE DIRECTIONS OF IMOTA
Testing
As mentioned, we had the ambition to enable first time users to work with IMOTA without requiring substantial time and at the same time to get concise results. Thus, we asked first-time users of IMOTA with a background either in biology or computer sciences to answer nine typical questions that we would expect users to ask (Supplemental Figure S3) as well as the ten questions from the System Usability Score (SUS) (https://www.usability.gov/how-to-and-tools/methods/system-usability-scale.html). We included on-site participants as well as remote users to apply IMOTA. The feedback was evaluated according to the guidelines of the U.S. Department of Health and Human Services (https://usability.gov). Twenty-three users participated in the study. The median time to complete the questions was 6 min and 53 s. The median success rate of users was 82% correct answers. Importantly, the final questions were almost without any exception correctly answered while errors have done almost exclusively in the first questions, highlighting that users get quickly familiar with IMOTA. The details on the success and error rates as well as execution times of test users are presented in Supplemental Figure S4. A median SUS of 85 was achieved (corresponding to an ‘excellent’ user experience). Given that users were non-experts with almost no background in the field and without training or getting background information we considered this already as very successful. At the same time, we learned from the failures and incorporated the feedback in the current version of IMOTA.
During the testing phase we evaluated different operating systems and browsers for the same queries as mentioned above. Independent of the operating system (Linux, MacOS, Windows) we observed no problems using Firefox or Chrome. Limited functionality of IMOTA may be found in using Internet Explorer and Safari, mostly due to the use of JavaScript. We, thus recommend using e.g. Firefox.
Limitations and future directions
(i) Currently, gene-, protein- and miRNA expression are included. Our aim is to further add on other omics types, most importantly epigenomics and metabolomics. (ii) With IMOTA we have a clear focus on Homo sapiens as organism. Although selected data are also available for other organisms, we currently do not plan to incorporate these in IMOTA but rather to add on to the content for H. sapiens. (iii) Only the miRNA tissue profiles have been measured from the same individuals. This is important to minimize inter-individual differences. For proteins and gene expression respective datasets will improve our understanding on organ specificity further. An important aspect that we will pursue is finally to provide autologous measurements, i.e. to include gene-, protein- and miRNA expression of organs from the same individuals to further minimize effects due to variations between different individuals. A respective analysis will also allow to correlate miRNA to gene and protein expression across different tissues directly, likely improving our understanding on how miRNAs target genes. (iv) In comparing and integrating high-throughput omics datasets there is always the risk to compare apples with oranges, especially if different platforms are used. In our case, the RNA expression data were measured by RNAseq while the miRNA data have been measured using microarrays. We thus will provide also NGS measurements for the miRNA data. (v) Another feature that will be added is the correlation of the genes, miRNAs and proteins to human pathologies.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Internal funds of Saarland University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Petryszak R., Keays M., Tang Y.A., Fonseca N.A., Barrera E., Burdett T., Fullgrabe A., Fuentes A.M., Jupp S., Koskinen S. et al. Expression Atlas update–an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016; 44:D746–D752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 3. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S. et al. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010; 28:1248–1250. [DOI] [PubMed] [Google Scholar]
- 4. Consortium G.T. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015; 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H. et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014; 509:582–587. [DOI] [PubMed] [Google Scholar]
- 6. Fehlmann T., Reinheimer S., Geng C., Su X., Drmanac S., Alexeev A., Zhang C., Backes C., Ludwig N., Hart M. et al. cPAS-based sequencing on the BGISEQ-500 to explore small non-coding RNAs. Clin. Epigenet. 2016; 8:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edgar R., Domrachev M., Lash A.E.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ienasescu H., Li K., Andersson R., Vitezic M., Rennie S., Chen Y., Vitting-Seerup K., Lagoni E., Boyd M., Bornholdt J. et al. On-the-fly selection of cell-specific enhancers, genes, miRNAs and proteins across the human body using SlideBase. Database (Oxford). 2016; 2016:baw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C.L., Haase J., Janes J., Huss J.W. III et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009; 10:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lizio M., Harshbarger J., Abugessaisa I., Noguchi S., Kondo A., Severin J., Mungall C., Arenillas D., Mathelier A., Medvedeva Y.A. et al. Update of the FANTOM web resource: high resolution transcriptome of diverse cell types in mammals. Nucleic Acids Res. 2017; 45:D737–D743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004; 32:D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsu S.D., Lin F.M., Wu W.Y., Liang C., Huang W.C., Chan W.L., Tsai W.T., Chen G.Z., Lee C.J., Chiu C.M. et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011; 39:D163–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Agarwal V., Bell G.W., Nam J.W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huber W., von Heydebreck A., Sultmann H., Poustka A., Vingron M.. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002; 18(Suppl. 1):S96–S104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.