Abstract
Here, we present the updated rSNPBase 3.0 database (http://rsnp3.psych.ac.cn), which provides human SNP-related regulatory elements, element-gene pairs and SNP-based regulatory networks. This database is the updated version of the SNP regulatory annotation database rSNPBase and rVarBase. In comparison to the last two versions, there are both structural and data adjustments in rSNPBase 3.0: (i) The most significant new feature is the expansion of analysis scope from SNP-related regulatory elements to include regulatory element–target gene pairs (E–G pairs), therefore it can provide SNP-based gene regulatory networks. (ii) Web function was modified according to data content and a new network search module is provided in the rSNPBase 3.0 in addition to the previous regulatory SNP (rSNP) search module. The two search modules support data query for detailed information (related-elements, element-gene pairs, and other extended annotations) on specific SNPs and SNP-related graphic networks constructed by interacting transcription factors (TFs), miRNAs and genes. (3) The type of regulatory elements was modified and enriched. To our best knowledge, the updated rSNPBase 3.0 is the first data tool supports SNP functional analysis from a regulatory network prospective, it will provide both a comprehensive understanding and concrete guidance for SNP-related regulatory studies.
INTRODUCTION
Biological and clinical studies that start from human genetic data, such as results of genome-wide association study (GWAS) or next-generation sequencing (NGS) study, largely depend on the subsequent functional analysis of detected SNPs (1,2). Considering the distribution ratio of coding and non-coding regions on the human chromosome as well as the increased evidence of the association between non-coding regions and a variety of human traits (3,4), the regulatory annotation of trait-associated SNPs has become more and more important in related studies. Therefore, we developed the rSNPBase (5) to provide a reliable regulatory annotation of human SNPs; we further expanded the annotation to include several types of known and novel human variants in the updated rVarBase (6). In the past four years, the two databases have been used in many disease genetic studies (7) and SNP functional studies (8).
While advances in human epigenetics enable researchers better understand complex disease molecular processes, new requirements for data analysis approaches and the corresponding data tools have appeared. An obviously trend in recent years is to use appropriate methods to gain a systematic view on interested traits (9). In such fields, gene regulatory network analysis is a useful method to understand the epigenetic mechanism underlying disease (10–12). One type of gene regulatory network is the transcription factor (TF) and microRNA (miRNA) co-regulatory network, which utilizes both transcriptional and post-transcriptional regulation relationships by integrating TF-gene and miRNA-gene interactions (13,14). This kind of analysis has been used in many disease molecular studies and reveals valuable mechanical evidence (15–17). There are several data tools to help users construct TF-miRNA co-regulatory networks by using disease expression data or disease-related elements as input (18–20), but to our best knowledge no data tool supports SNP functional analysis from a regulatory network prospective. To facilitate researchers to perform gene regulatory network analysis based on genetic results, we made both structural and data adjustments to the rSNPBase and present it as the updated version rSNPBase 3.0 (http:// rsnp3.psych.ac.cn).
In rSNPBase 3.0, human genome SNPs were annotated with both related regulatory elements and regulatory element-target gene pairs (E–G pairs). In addition to the regulatory SNP (rSNP) search module that provides detailed epigenetic features for queried SNPs and their linkage disequilibrium (LD) proxies, the database also provides a network analysis module, which can generate gene regulatory networks to allow users to get a more intuitive understanding of regulatory elements, processes, and mechanisms related to their queried variants. Nodes in the generated network represent regulatory elements (TF and miRNA) and target genes, and edges between nodes show SNPs that are related to the specific E–G pairs. The combination of the two modules will help users to get an overall logical understanding of their trait of interest as well as get more concrete guidance for follow-up studies.
DATA CONTENT AND NEW FEATURES
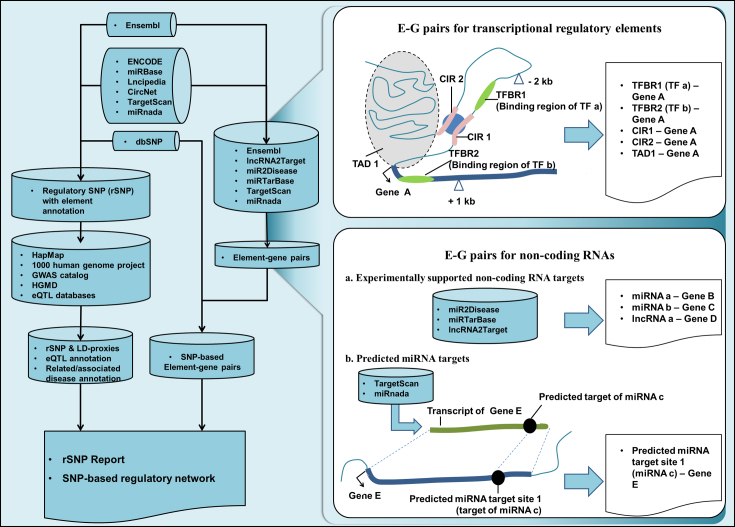
Beyond adding newly created small variations from the NCBI dbSNP database, new types of regulatory elements and more extended annotations (such as SNP-related diseases), a significant adjustment in the database structure and function have been made in this update. The concerns of rSNPBase 3.0 extended from regulatory elements that overlapped SNPs to SNP-related E–G pairs. Based on these changes, the updated database supports SNP-based gene regulatory network analysis. As shown in Figure 1, seven types of regulatory elements were derived from reference databases (most of them provide experimental supported data), and the relationship between the included regulatory elements and genes from Ensembl (GRCh37) (21) were analysed by genomic proximity or by using reference databases. Genome-wide human SNPs from NCBI dbSNP (build 150) (22) were analysed and filtered with involved regulatory elements, and were therefore connected to corresponding E–G pairs at the same time. The analysis results were stored in rSNPBase 3.0 and presented as rSNP reports and SNP-based regulatory networks. A summarized data contents of rSNPBase 3.0 is shown in Table 1. Detailed information about the utilized regulatory elements and reference database used in the data processing is shown in the supplementary data and at http://rsnp3.psych.ac.cn/datacontent.do.
Figure 1.
Data processing of rSNPBase 3.0.
Table 1. Data content of rSNPBase 3.0 (as of 15 September 2017) and comparison of the current and previous versions.
| Involved SNPs in three version | rSNPBase | rVarBase | rSNPBase 3.0 |
|---|---|---|---|
| rSNPs | 22 846 898 | 87 345 304 | 117 452 549 |
| LD-proxies of rSNPs (non-rSNPs) | 2 281 874 | 1 626 737 | 2 096 231 |
| SNP related regulatory elements | |||
| Transcription factor binding regions (TFBRs) | 7 562 592 | ||
| Chromatin interactive regions (CIRs) | 212 837 | ||
| Topologically associated domains (TADs) | 38 916 | ||
| Mature microRNA (miRNA) regions | 2 794 | ||
| Predicted miRNA target sites | 384 284 | ||
| Long non-coding RNA (lncRNA) regions | 211 749 | ||
| Circular RNA (circRNA) regions | 312 673 | ||
| SNP related regulatory element-target gene (E–G) pairs | |||
| TFBR-gene pairs | 9 484 132 | ||
| CIR - gene pairs | 321 247 | ||
| TAD – gene pairs | 48 482 | ||
| miRNA - gene pairs | |||
| Experimentally supported | 37 781 | ||
| Predicted | 384 284 | ||
| lncRNA - gene pairs | 4 092 | ||
| Extend annotations | |||
| Diseases(SNP-disease pairs) | 97 896 | ||
| eQTLs (SNP-mRNA pairs) | 4 201 218 | ||
Regulatory elements involved in rSNPBase 3.0
rSNPBase 3.0 is consistent with previous versions in annotating SNPs with reliable regulatory elements (mainly identified by experiments). There are seven types of regulatory elements included. Experimentally identified transcription factor binding regions (TFBRs) and chromatin interactive regions (CIRs) from the Encyclopaedia of DNA Elements (ENCODE) project (23), validated mature miRNA regions from miRBase (24), predicted human miRNA target sites in 3′-UTR regions from TargetScan (25,26) and miRNAda (27), and validated long non-coding RNAs (lncRNAs) region from LNCipedia (28) were included in the two previous versions and remain in this update. One of the greatest progress in human epigenetic in recent years is advances in 3D chromatin structures (29,30), which expands the research dimensions from DNA accessibility and DNA binding to long-distance element interactions. Thus, in rVarBase 3.0, we consolidated the topologically associated domains (TADs) from ENCODE-processed data. Circular RNAs (circRNAs) from CircNet were also added (31), along with accumulation information for this type of non-coding RNA. Although they contain rich and important information, DNA elements that describe epigenetic statues on large chromosome scales, such as active chromatin state regions, histone binding regions, and methylation of CpG islands may suffer a limited impact from the genotype changes of small variations, so they were not included in rSNPBase 3.0.
Regulatory elements-gene pairs (E–G pairs) and gene regulatory network
Except for circRNAs, target gene analyses were performed on the other six types of regulatory elements to get corresponding E–G pairs.
E–G pairs for transcriptional regulatory elements
Chromosome location of transcriptional regulatory elements was acquired from ENCODE Consortium, among which TFBRs information was acquired from proceeded ChIP-seq peak data, CIRs information was from Chromatin Interactions by 5C and ChIA-PET, and TADs information was from proceeded Hi-C data. Genome locations of the three types of regulatory elements were compared with the potential promoter region (from 2k upstream to 1k downstream of transcription start sites) of all Ensembl recorded genes on hg19 coordinate. As illustrated in Figure 1, if TFBRs (TFBR1 and TFBR2 in the illustration) were located in the potential promoter region of a gene (Gene A), the E–G pairs of TFBR1-Gene A and TFBR2-Gene A were constructed. The connection between TFs that binding to the TFBRs (TF a and TF b in the illustration) and Gene A were constructed correspondingly to support subsequently TF-gene network analysis. For CIRs, if one fragment (CIR1) in the interactive regions was located in potential promoter region of the Gene A, the relationships between both the interacted DNA fragments (CIR1 and CIR2) and Gene A were constructed. TADs were also connected to genes with overlapped chromosome location.
E–G pairs for non-coding RNAs
The regulatory targets of miRNAs and lncRNAs have been accumulated using experimental data and prediction methods. The databases of miR2Disease (32), miRTarBase (33) and lncRNA2Target (34) include experimentally supported target relationships between miRNAs/lncRNAs and genes that validated by RT-PCR, microarray or RNA-seq. These data were integrated into rSNPBase 3.0 as miRNA–gene pairs and lncRNA-gene pairs directly. The databases of TargetScan (25,26) and miRnada (27) include predicted miRNA target sites in transcripts of several species. Human transcripts and predicted targets of human miRNAs were obtained and mapped to human genome sequence (on hg 19 coordinate). As shown in Figure 1, chromosome location of genome sequence corresponding to predicted target site in transcript (Predicted miRNA target site 1) and gene that coding the target transcript (Gene E) were added into the E–G pair list. The miRNA (miRNA c in the illustration) that was predicted to match to the site was also connected with Gene E to support subsequently miRNA-gene network analysis.
SNP-based E–G pairs and gene regulatory network
All the six types of E–G pairs were used to annotate SNPs by comparing the chromosomal location between the SNP and the element in E–G pair. SNP-related E–G pairs were stored and could be used as a basic unit to build an SNP-related regulatory network. In addition to allowing researchers to browse E–G pairs as part of the SNP annotation information, rSNPBase 3.0 provides specific module for showing graphical gene regulatory networks that have been constructed based on SNP-related TFBR-gene pairs, mature miRNA region-gene pairs and predicted miRNA target site-gene pairs. Nodes in the network represent corresponding TFs, miRNAs, and genes.
Extended information
Three types of extended information were added to the rSNPs. To cover genetic-correlated SNPs, LD correlations between human SNPs were analysed according to the 1000 Genomes project (35) and HapMap (36). Non-rSNPs with strong LD (r2>0.8) with rSNPs were added as LD-proxies. To provide more functional evidence, expression quantitative trait loci (eQTLs) information was annotated for all the included SNPs (both rSNPs and their LD-proxies) with references to several experimentally support databases (37,38). To facilitate pathogenicity studies, SNP-associated diseases were also annotated. In addition to the associations between SNPs and complex diseases from the GWAS catalogue (39), relationships between SNPs and inherited diseases from the Human Gene Mutation Database (HGMD) (40) were also included in rSNPBase 3.0.
WEB INTERFACE AND DATABASE USAGE
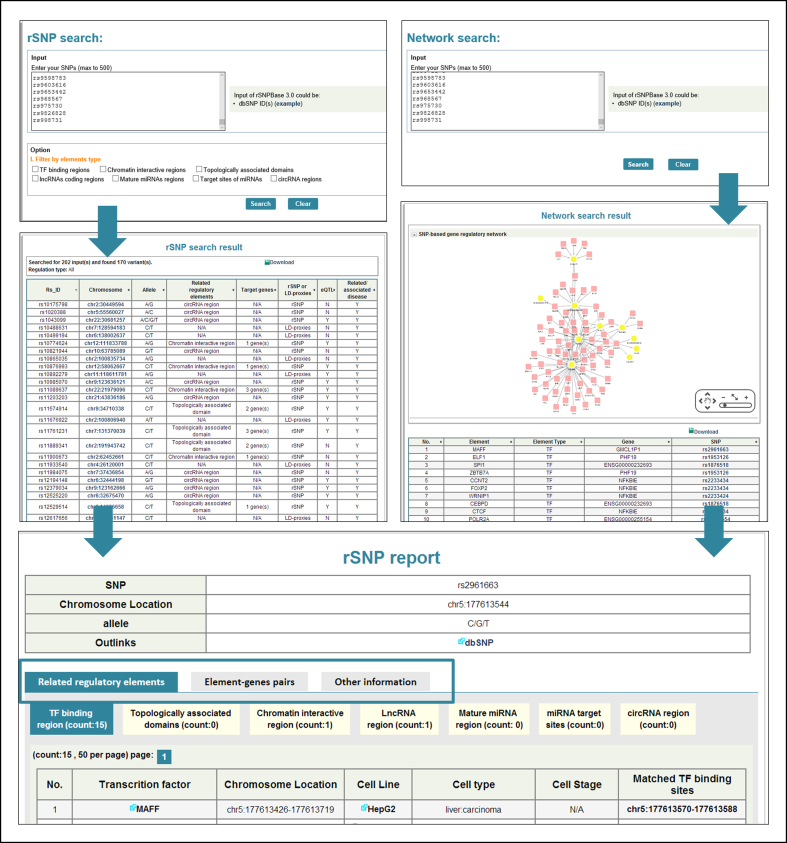
The web interface for rSNPBase 3.0 was constructed with the following two main function modules: ‘rSNP search’ and ‘Network search’. As shown in Figure 2, the ‘rSNP search’ module contains almost all the functions in the previous version. Users could query SNPs by using dbSNP rs IDs and then get summarized regulatory features and extended information on the queried SNPs in the ‘Search Results’ page. The search results could be downloaded and a detailed SNP information report could be seen in the linked ‘SNP Report’ page. The new ‘Network search’ module, which is shown in Figure 2, can also use dbSNP rs IDs as input data. The data query in this module results in a graphic regulatory network and an ‘element-gene-related SNPs’ interaction table, which can be downloaded and modulated with graphic tools such as Cytoscape (41). In addition to the two search modules, an FTP site was provided to facilitate the download of annotations for all SNPs included in the database.
Figure 2.
Web interface and data retrieving process of rSNPBase 3.0.
In addition to providing a potential functional interpretation for individual results from genetic studies, the updated rSNPBase 3.0 may support deeper and more comprehensive functional inferences. Here, we present database usage with a demo SNP set. The demo set includes 199 SNPs that were recorded in GWAS catalogue as rheumatoid arthritis (RA)-associated SNPs (P < 1E–5 in GWAS), as shown in the supplementary data. Similar to the previous version, the regulatory features of the demo SNPs could be directly searched in the ‘rSNP search’ module. As shown in Figure 2, the search results show that 167 of the 199 SNPs are included in rSNPBase 3.0. In total, 115 of the included SNPs are rSNPs that are related to TFBRs, TADs, CIRs, lncRNAs and circRNAs; the other 52 are LD-proxies of rSNPs. The demo SNPs could also be searched in the new ‘Network search’ module and generated a network constructed by 81 nodes and 115 edges. The 81 nodes represent 71 TFs and 10 genes, and the NFKBIE gene, PHF19 gene and the TF POLR2A occupy the most central positions in the network. This network illustrates potential evidence for gene regulation related to RA-associated SNPs, and may provide comprehensive information for subsequent functional analyses and experiments. For a specific TF-gene pair, detailed information could be observed in the rSNP report page using a link from the SNP in the ‘element-gene-related SNPs’ interaction table below the graphical network. The combination of systematic prospects and detailed regulation information will provide practical references for functional studies with genetic results.
CONCLUSIONS AND FUTURE PLAN
Here, we present rSNPBase 3.0, an updated SNP regulatory database. It extends the annotation scope of SNP-related regulatory elements to SNP-related regulatory element-target gene pairs and can therefore support SNP-based gene regulatory network analysis. In addition to providing intuitive SNP regulatory annotations that could be used as concrete guidance for follow-up studies, the updated rSNPBase 3.0 also allows researchers to get a more systematic view of the potential regulatory mechanisms related to their SNPs of interest. Both SNP-related E–G pairs and SNP-based TF/microRNA co-regulatory networks will provide logical evidence that bridges genetic results with specific complex disease studies. In addition to improving the data content, the web interface was also modified to a more concise and user-friendly manner.
rSNPBase 3.0 will be continuously updated. In addition to adding newly reported human genetics and epigenetics data, more comprehensive analysis approaches will be considered. For example, the gene regulatory networks are connected and interplay with other types of interactive networks, such as signal transduction networks (42), protein–protein interaction networks (43), and protein phosphorylation networks (44). We will pay close attention to the frontier data and methods in this field, and aim to integrate this information in an appropriate and timely manner.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
CAS Key Laboratory of Mental Health, Institute of Psychology; CAS/SAFEA International Partnership Program for Creative Research Teams [Y2CX131003]; Knowledge Innovation Program of the Chinese Academy of Sciences [KSCX2-EW-J-8]; National Natural Science Foundation of China [81201046]. Funding for open access charge: CAS Key Laboratory of Mental Health, Institute of Psychology; CAS/SAFEA International Partnership Program for Creative Research Teams [Y2CX131003]; Knowledge Innovation Program of the Chinese Academy of Sciences [KSCX2-EW-J-8]; National Natural Science Foundation of China [81201046].
Conflict of interest statement. None declared.
REFERENCES
- 1. Edwards S.L., Beesley J., French J.D., Dunning A.M.. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013; 93:779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nishizaki S.S., Boyle A.P.. Mining the Unknown: Assigning Function to Noncoding Single Nucleotide Polymorphisms. Trends Genet. 2017; 33:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Devanna P., Chen X.S., Ho J., Gajewski D., Smith S.D., Gialluisi A., Francks C., Fisher S.E., Newbury D.F., Vernes S.C.. Next-gen sequencing identifies non-coding variation disrupting miRNA-binding sites in neurological disorders. Mol. Psychiatry. 2017; doi:10.1038/mp.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Khurana E., Fu Y., Chakravarty D., Demichelis F., Rubin M.A., Gerstein M.. Role of non-coding sequence variants in cancer. Nat. Rev. Genet. 2016; 17:93–108. [DOI] [PubMed] [Google Scholar]
- 5. Guo L., Du Y., Chang S., Zhang K., Wang J.. rSNPBase: a database for curated regulatory SNPs. Nucleic Acids Res. 2014; 42:D1033–D1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guo L., Du Y., Qu S., Wang J.. rVarBase: an updated database for regulatory features of human variants. Nucleic Acids Res. 2016; 44:D888–D893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun C., Molineros J.E., Looger L.L., Zhou X.J., Kim K., Okada Y., Ma J., Qi Y.Y., Kim-Howard X., Motghare P. et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat. Genet. 2016; 48:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arkova O.V., Ponomarenko M.P., Rasskazov D.A., Drachkova I.A., Arshinova T.V., Ponomarenko P.M., Savinkova L.K., Kolchanov N.A.. Obesity-related known and candidate SNP markers can significantly change affinity of TATA-binding protein for human gene promoters. BMC Genomics. 2015; 16:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koch L. Genetics: From non-coding risk variant to biological mechanism in CAD. Nat. Rev. Cardiol. 2017; 14:569. [DOI] [PubMed] [Google Scholar]
- 10. Emmert-Streib F., Dehmer M., Haibe-Kains B.. Gene regulatory networks and their applications: understanding biological and medical problems in terms of networks. Front. Cell Dev. Biol. 2014; 2:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Emmert-Streib F., de Matos Simoes R., Mullan P., Haibe-Kains B., Dehmer M.. The gene regulatory network for breast cancer: integrated regulatory landscape of cancer hallmarks. Front Genet. 2014; 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singh H., Khan A.A., Dinner A.R.. Gene regulatory networks in the immune system. Trends Immunol. 2014; 35:211–218. [DOI] [PubMed] [Google Scholar]
- 13. Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008; 319:1785–1786. [DOI] [PubMed] [Google Scholar]
- 14. Guo A.Y., Sun J., Jia P., Zhao Z.. A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC Syst. Biol. 2010; 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ye H., Liu X., Lv M., Wu Y., Kuang S., Gong J., Yuan P., Zhong Z., Li Q., Jia H. et al. MicroRNA and transcription factor co-regulatory network analysis reveals miR-19 inhibits CYLD in T-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2012; 40:5201–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hsieh W.T., Tzeng K.R., Ciou J.S., Tsai J.J., Kurubanjerdjit N., Huang C.H., Ng K.L.. Transcription factor and microRNA-regulated network motifs for cancer and signal transduction networks. BMC Syst. Biol. 2015; 9:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poos K., Smida J., Nathrath M., Maugg D., Baumhoer D., Korsching E.. How microRNA and transcription factor co-regulatory networks affect osteosarcoma cell proliferation. PLoS Comput. Biol. 2013; 9:e1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamed M., Spaniol C., Nazarieh M., Helms V.. TFmiR: a web server for constructing and analyzing disease-specific transcription factor and miRNA co-regulatory networks. Nucleic Acids Res. 2015; 43:W283–W288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vlachos I.S., Vergoulis T., Paraskevopoulou M.D., Lykokanellos F., Georgakilas G., Georgiou P., Chatzopoulos S., Karagkouni D., Christodoulou F., Dalamagas T. et al. DIANA-mirExTra v2.0: Uncovering microRNAs and transcription factors with crucial roles in NGS expression data. Nucleic Acids Res. 2016; 44:W128–W134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Z.P., Wu C., Miao H., Wu H.. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database (Oxford). 2015; bav095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aken B.L., Ayling S., Barrell D., Clarke L., Curwen V., Fairley S., Fernandez Banet J., Billis K., Garcia Giron C., Hourlier T. et al. The Ensembl gene annotation system. Database (Oxford). 2016; baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T. et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016; 44:D726–D732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006; 342:129–138. [DOI] [PubMed] [Google Scholar]
- 25. Lewis B.P., Burge C.B., Bartel D.P.. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120:15–20. [DOI] [PubMed] [Google Scholar]
- 26. Friedman R.C., Farh K.K., Burge C.B., Bartel D.P.. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Betel D., Wilson M., Gabow A., Marks D.S., Sander C.. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008; 36:D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P.. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015; 43:D174–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmitt A.D., Hu M., Ren B.. Genome-wide mapping and analysis of chromosome architecture. Nat. Rev. Mol. Cell Biol. 2016; 17:743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krijger P.H., de Laat W.. Regulation of disease-associated gene expression in the 3D genome. Nat. Rev. Mol. Cell Biol. 2016; 17:771–782. [DOI] [PubMed] [Google Scholar]
- 31. Liu Y.C., Li J.R., Sun C.H., Andrews E., Chao R.F., Lin F.M., Weng S.L., Hsu S.D., Huang C.C., Cheng C. et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016; 44:D209–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y.. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009; 37:D98–D104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chou C.H., Chang N.W., Shrestha S., Hsu S.D., Lin Y.L., Lee W.H., Yang C.D., Hong H.C., Wei T.Y., Tu S.J. et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016; 44:D239–D247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang Q., Wang J., Wu X., Ma R., Zhang T., Jin S., Han Z., Tan R., Peng J., Liu G. et al. LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res. 2015; 43:D193–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Genomes Project, C. Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A.. A map of human genome variation from population-scale sequencing. Nature. 2010; 467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R.. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010; 34:816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Consortium, G.T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015; 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma B., Huang J., Liang L.. RTeQTL: Real-Time Online Engine for Expression Quantitative Trait Loci Analyses. Database (Oxford). 2014; bau066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shaw K., Cooper D.N.. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr. Protoc. Bioinformatics. 2012; doi:10.1002/0471250953.bi0113s39. [DOI] [PubMed] [Google Scholar]
- 41. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goncalves E., Bucher J., Ryll A., Niklas J., Mauch K., Klamt S., Rocha M., Saez-Rodriguez J.. Bridging the layers: towards integration of signal transduction, regulation and metabolism into mathematical models. Mol. Biosyst. 2013; 9:1576–1583. [DOI] [PubMed] [Google Scholar]
- 43. Padi M., Quackenbush J.. Integrating transcriptional and protein interaction networks to prioritize condition-specific master regulators. BMC Syst. Biol. 2015; 9:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Newman R.H., Zhang J., Zhu H.. Toward a systems-level view of dynamic phosphorylation networks. Front. Genet. 2014; 5:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.