Abstract
As a traditional medical intervention in Asia and a complementary and alternative medicine in western countries, Traditional Chinese Medicine (TCM) is capturing worldwide attention in life science field. Traditional Chinese Medicine Integrated Database (TCMID), which was originally launched in 2013, was a comprehensive database aiming at TCM’s modernization and standardization. It has been highly recognized among pharmacologists and scholars in TCM researches. The latest release, TCMID 2.0 (http://www.megabionet.org/tcmid/), replenished the preceding database with 18 203 herbal ingredients, 15 prescriptions, 82 related targets, 1356 drugs, 842 diseases and numerous new connections between them. Considering that chemical changes might take place in decocting process of prescriptions, which may result in new ingredients, new data containing the prescription ingredients was collected in current version. In addition, 778 herbal mass spectrometry (MS) spectra related to 170 herbs were appended to show the variation of herbal quality in different origin and distinguish genuine medicinal materials from common ones while 3895 MS spectra of 729 ingredients were added as the supplementary materials of component identification. With the significant increase of data, TCMID 2.0 will further facilitate TCM’s modernization and enhance the exploration of underlying biological processes that are response to the diverse pharmacologic actions of TCM.
INTRODUCTION
In the past, most of research achievements in prescriptions, herbs, ingredients and other Traditional Chinese Medicine (TCM) related information were dispersedly recorded in books and journals, which had hampered systematical investigations and applications for TCM. However, this situation began to change when Traditional Chinese Medicine Integrated Database (TCMID) and other TCM related databases were launched. Comprehensive integration of various data resources, including Database@Taiwan (1), HIT (2) makes TCMID as the largest database of TCM in related field with over 49 000 prescriptions, 8159 herbs, 25 210 ingredients, 3791 diseases, 6828 drugs and 17 521 targets (3). Links with open access databases, such as Drugbank (4), OMIM (5) and STITCH (6) are available in TCMID. These links provide additional detailed descriptions about relevant drugs, diseases and targets. TCMID has attracted considerable visit volume from researchers of related field in the past 4 years and has significantly promoted mechanism studies in prescriptions and herbs through multi-level approaches (7,8), making an advance in bioinformatics research of TCM (9).
In recent years, attention to TCM continue to grow, attracting academic circles for the extensive research in therapeutic herbal ingredients, such as baicalein that was suggested as a pharmacotherapies for abdominal aortic aneurysm (10) and neurodegenerative disease (11) and ginsenoside, which was applied in various cardiovascular diseases (12). Great efforts have been made to extract and isolate effective chemical in herbs and prescriptions over the past 4 years, resulting in emergence of quantity of data about newly identified ingredients. As a consequence, data in databases largely lag behind factual status. Information about ingredients is the fundamental element in various types of TCM researches. Incompletely collected information may lead to the deviation of accuracy in the systematic researches that attempt to unveil the therapeutic mechanism of TCM.
China is vast in territory and the majority of herbs have more than one place of origin. Herbs in different districts and growth condition vary in effective composition content that is responsible for the otherness in curative effect (13). Lack of stable quality control in herbs still blocks the way of TCM modernization. Recently, researchers have come to realize the urgency to regulate quality control and have made great efforts to solve this problem initiatively (14,15). On account of such endeavour, many mass spectrometry (MS) spectra are now available. However, no TCM related databases has compiled such kind of data yet.
Therefore, a database with more comprehensive and accurate recording related data and information is in demand. In updated TCMID 2.0, the original data is largely expanded while two new data fields, prescription ingredients and MS spectra, have been added. This new release will be a more useful resource for systemic TCM researches and further promote the TCM applications worldwide. New configuration of the web interface is now available at http://www.megabionet.org/tcmid/.
RESULTS
Data updates and extensions
Presentation of TCMID 2.0, including three relatively independent sections, is described as follows (Table 1). The first section primarily focuses on the enhancements made to the original part. In this section, 18 203 herbal ingredients were manually collected through literature mining and were then compiled into the database which largely extended the data of this part. More links of chemical–target, target–drug and target–disease were generated which brought 82 new targets, 1356 new drugs, 842 diseases into the database. Moreover, 15 new prescriptions were found during the literature surveys and 176 315 protein–protein interaction (PPI) pairs were integrated. In the second section, we introduced the augment of extra data fields–prescription ingredients and MS spectra of herbs and ingredients. Relevant information of this section was assigned to 1072 prescriptions, 170 herbs or 729 ingredients respectively. The last section is about configuration of the latest redesigned webpage of the database.
Table 1. Overview of the number of data in TCMID and TCMID 2.0.
| NO. of original data | No. of present data | |
|---|---|---|
| Prescriptions | 46 914 | 46 929 |
| Herbs | 8159 | 8159 |
| Total ingredients | 25 210 | 43 413 |
| Drugs | 6826 | 8182 |
| Diseases | 3791 | 4633 |
| Prescription ingredients | 0 | 1045 |
| Herbal mass spectra | 0 | 778 |
| Mass spectrometry of ingredients | 0 | 3895 |
Enhancements of original field
Compared with the former version, TCMID 2.0 was greatly enhanced by extensive data mined from published literature and integrated from other available open resources. TCMID encompassed six mutually connected parts, prescriptions, herbs, ingredients, diseases, drugs and targets with detailed descriptions and information. Since most studies working on the separation of active components in herbs were published in Chinese, we primarily utilized Chinese national knowledge infrastructure (CNKI) (http://www.cnki.net/) to collect related literature information in regard to those 8159 recorded herbs in TCMID. After the endeavour on literature mining, we appended 18 203 variant ingredients for 639 herbs, accounting for a 72.2% increase in the count of previous herbal ingredients. Details about the ingredients, including SMILE string, formula and 2D structure were extracted from PubChem (16). ChEBI (http://www.ebi.ac.uk/chebi) (17) and UNPD (http://unpd.chem960.com/) were applied to help us map those chemicals whose names were not available in PubChem. Chemical-protein interactions in Homo sapiens were acquired from STITCH 5.0 (http://stitch.embl.de/) (6). Moreover, we redesigned the target table in ingredient page which convey more valuable information about each target including the confidence score from STITCH, association mode, detailed action between ingredients and targets, ENSP_ID and UniProt_ID. This information can give users a better understanding of the relationship between herbal ingredients and their targets. Through this step, 82 new targets were obtained. 176 315 PPIs were acquired for the targets in database with the latest data in BioGrid database (18). Additionally, 15 new prescriptions were identified during the literature search. These 15 new prescriptions and their compositions were then recorded into the latest database.
Above-mentioned efforts make our database keep pace with the latest relevant research achievements, which will facilitate the systematical researches in TCM.
Additions of new data fields
Two completely new data fields were integrated to TCMID 2.0, namely the prescription ingredients and MS spectra. Since prescriptions commonly consist of more than one herb and every herb contains various ingredients, chemical reaction may also take place during the decoction process of the mixture and would sequentially cause alteration in chemical extraction characteristics (19). Therefore, conventional bioinformatics approaches solely based on ingredients of herbs may not be able to fully decipher the molecular mechanism of prescriptions. Taking these factors into consideration, we added a new data field containing the prescription ingredients. Similarly, CNKI were applied for collecting literature data. Finally, 1072 prescriptions with 897 extracted ingredients were collected through the literature. Among them, 365 of the extracted ingredients were not matched to available herbal ingredients, confirming that the ingredients of a certain prescription were not completely equal to a simple summarization of ingredients of each herb. This finding clarified the necessity and importance of the new data field.
Another field is MS spectra. Mass spectrometry is a mature technique to discern characterization of substances. To address the predicament in quality control of Chinese herbs, we gathered the information of herbal MS spectra for the recorded herbs in TCMID through retrieving the CNKI. Each MS spectrum was labelled with the origin place of an herb, types of chromatography and extracted chemical compounds with their appearing locations. A standard MS spectrum was always placed along with the common one. Ultimately, after manual collection and screening, we obtained 778 mass spectra for 170 different herbs such as Chuan Xin Lian (Andrographis paniculata [Syn. Justicia paniculata]). In addition, 3895 raw MS spectra, corresponding to 729 ingredients, were retrieved from GNSP, a freely available database offering retrieval and reanalysis of MS spectra for natural products (20).
Enhanced querying and viewing capabilities
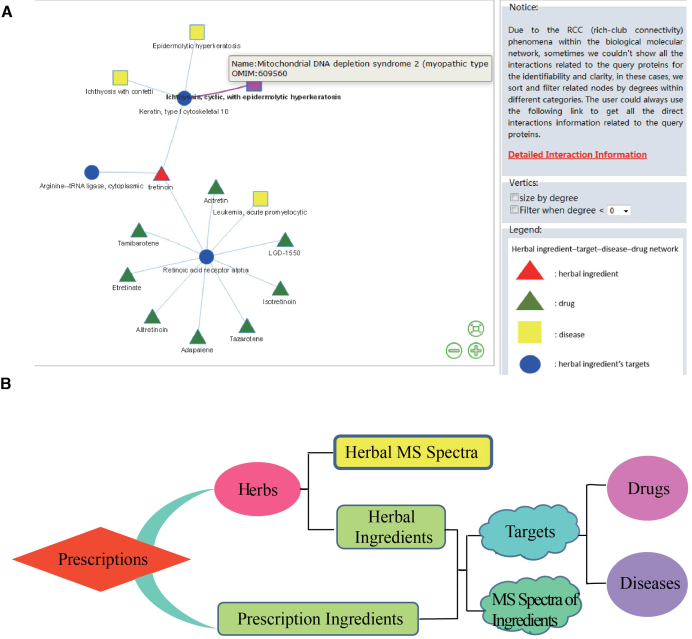
To offer a user more friendly interface, we updated the webpage. Six original data fields used to query the database are still reachable. Besides four newly added tables in prescriptions (ingredients), herbs (MS spectra), ingredients (MS spectra) and targets (PPIs), we reprogrammed the network display tool by replacing the Java Applet with vis.js, the dynamic and browser based visualization library. In some case, we sorted and filtered the nodes by degree since some networks carrying all the interactions related to the query node may cause visual clutter. Links carrying complete data of each network are available in the tooltip of the view page. To realize maximal display and optimize web data transmission, online display tools were redeveloped and detailed information of each node was annotated in the display page (Figure 1A).
Figure 1.
Data display and structure of TCMID 2.0. (A) An herbal ingredient-target network as an illustration for the new visualizations of networks. The red triangle stands for the ingredient used for query; the blue circle stands for targets of ingredients; the green triangle stands for the herbal targets related drugs; the yellow square stands for the herbal targets related diseases. Information text of nodes is reachable when users drag the mouse over one node and the colour of the node will turn to purple. Direct interaction information related to the query node can be viewed through the link in the right tooltip. (B) An illustration of internal connections among each data piece.
CONCLUSIONS
Over the past 4 years, TCMID has shown its increasing importance as a connector to link TCM with modern researches. As highlight throughout the paper, TCMID 2.0 endeavours in making a more comprehensive and integrated database of TCM to accelerate the progress of TCM’s modernization and standardization. It has made great enhancements and updates over TCMID while almost retained the original network structure (Figure 1B). We believe that TCMID 2.0 can provide richer information and better services to the related research community for more comprehensive studies in TCM.
FUNDING
National Natural Science Foundation of China [81373633 to C.W.]; National High Technology Research and Development Program of China [2015AA020108 to T.S.]; Special Scientific Project of Traditional Chinese Medicine [201507001-4 to C.W.]; Major Special Science and Technology Project of Zhejiang Province [2014C03046-1 to C.W.]; 111 Project [B14019 to T.S.]. Funding for open access charge: National Natural Science Foundation of China [81373633 to C.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chen C.Y. TCM Database@Taiwan: the world's largest traditional Chinese medicine database for drug screening in silico. PLoS One. 2011; 6:e15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ye H., Ye L., Kang H., Zhang D., Tao L., Tang K., Liu X., Zhu R., Liu Q., Chen Y.Z. et al. HIT: linking herbal active ingredients to targets. Nucleic Acids Res. 2011; 39:D1055–D1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xue R., Fang Z., Zhang M., Yi Z., Wen C., Shi T.. TCMID: Traditional Chinese Medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res. 2013; 41:D1089–D1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knox C., Law V., Jewison T., Liu P., Ly S., Frolkis A., Pon A., Banco K., Mak C., Neveu V. et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011; 39:D1035–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baxevanis A.D. Searching Online Mendelian Inheritance in Man (OMIM) for information on genetic loci involved in human disease. Curr. Protoc. Hum. Genet. 2012; 13:11–10. [DOI] [PubMed] [Google Scholar]
- 6. Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P., Kuhn M.. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016; 44:D380–D384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang L., Lv Q., Liu F., Shi T., Wen C.. A Systems Biology-Based Investigation into the Pharmacological Mechanisms of Sheng-ma-bie-jia-tang Acting on Systemic Lupus Erythematosus by Multi-Level Data Integration. Sci. Rep. 2015; 5:16401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang L., Lv Q., Xie D., Shi T., Wen C.. Deciphering the potential pharmaceutical mechanism of Chinese Traditional Medicine (Gui-Zhi-Shao-Yao-Zhi-Mu) on rheumatoid arthritis. Sci. Rep. 2016; 6:22602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Z., Guo F., Wang Y., Li C., Zhang X., Li H., Diao L., Gu J., Wang W., Li D. et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechANism of Traditional Chinese Medicine. Sci. Rep. 2016; 6:21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang F., Chen H., Yan Y., Liu Y., Zhang S., Liu D.. Baicalein protects against the development of angiotensin II-induced abdominal aortic aneurysms by blocking JNK and p38 MAPK signaling. Science China. Life Sci. 2016; 59:940–949. [DOI] [PubMed] [Google Scholar]
- 11. Jiang W., Li S., Li X.. Therapeutic potential of berberine against neurodegenerative diseases. Sci. China. Life Sci. 2015; 58:564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun Y., Liu Y., Chen K.. Roles and mechanisms of ginsenoside in cardiovascular diseases: progress and perspectives. Sci. China. Life Sci. 2016; 59:292–298. [DOI] [PubMed] [Google Scholar]
- 13. Zhen G., Zhang L., Du Y., Yu R., Liu X., Cao F., Chang Q., Deng X., Xia M., He H.. De novo assembly and comparative analysis of root transcriptomes from different varieties of Panax ginseng C.A. Meyer grown in different environments. Sci. China. Life Sci. 2015; 58:1099–1110. [DOI] [PubMed] [Google Scholar]
- 14. Gao H., Wang Z., Li Y., Qian Z.. Overview of the quality standard research of traditional Chinese medicine. Front. Med. 2011; 5:195–202. [DOI] [PubMed] [Google Scholar]
- 15. Melchart D., Hager S., Dai J., Weidenhammer W.. Quality control and complication screening programme of Chinese medicinal drugs at the First German Hospital of Traditional Chinese Medicine—a retrospective analysis. Forsch. Komplementmed. 2016; 23(Suppl. 2):21–28. [DOI] [PubMed] [Google Scholar]
- 16. Coordinators N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013; 41:D8–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C.. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016; 44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chatr-Aryamontri A., Breitkreutz B.J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O’Donnell L. et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015; 43:D470–D478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim J.H., Ha W.R., Park J.H., Lee G., Choi G., Lee S.H., Kim Y.S.. Influence of herbal combinations on the extraction efficiencies of chemical compounds from Cinnamomum cassia, Paeonia lactiflora, and Glycyrrhiza uralensis, the herbal components of Gyeji-tang, evaluated by HPLC method. J. Pharm. Biomed. Anal. 2016; 129:50–59. [DOI] [PubMed] [Google Scholar]
- 20. Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C.A., Luzzatto-Knaan T. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016; 34:828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]