Abstract
MODOMICS is a database of RNA modifications that provides comprehensive information concerning the chemical structures of modified ribonucleosides, their biosynthetic pathways, the location of modified residues in RNA sequences, and RNA-modifying enzymes. In the current database version, we included the following new features and data: extended mass spectrometry and liquid chromatography data for modified nucleosides; links between human tRNA sequences and MINTbase - a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments; new, machine-friendly system of unified abbreviations for modified nucleoside names; sets of modified tRNA sequences for two bacterial species, updated collection of mammalian tRNA modifications, 19 newly identified modified ribonucleosides and 66 functionally characterized proteins involved in RNA modification. Data from MODOMICS have been linked to the RNAcentral database of RNA sequences. MODOMICS is available at http://modomics.genesilico.pl.
INTRODUCTION
The presence of modified nucleosides in RNA, beyond the basic A, U, C and G, has been recognized for more than half a century. However, their importance to RNA biochemistry and cell biology has been underappreciated, mainly because very few modifications (at defined positions in defined RNA molecules) were found to be truly indispensable for basic biological processes [review: (1)]. Now we know that 163 post-transcriptional modifications of RNA introduce a functional diversity that allows the four basic ribonucleotide residues to gain diverse functions, akin to those of side chains of amino acid residues, which may be e.g., polar, charged, aliphatic or aromatic. Modifications can directly influence RNA structure, by promoting or disrupting certain intramolecular interactions; they can make the RNA molecule more rigid or more flexible. They can also influence RNA interactions with other molecules, in particular proteins. Overall, they contribute strongly to the diversity of functions fulfilled by RNA molecules, especially within complex regulatory networks, where small subtle structural changes can bring about significant changes to cellular metabolism (2).
In the last years, new types of RNA modifications have been found, and biochemical and physiological roles have been elucidated for many known modified ribonucleosides (3–5). Some of these advances were driven by the use of liquid chromatography/mass spectrometry (LC/MS)-based methods, which provide highly precise quantification of changes in the spectrum of modified ribonucleosides in RNA from any organism, facilitating the study of translational control of cellular responses and phenotypes (6). Moreover, a number of previously unknown RNA-modifying enzymes have been identified and characterized.
New important roles for long-known RNA modifications were also discovered. Prominent examples include the involvement of N6-methyladenosine (m6A) in regulating gene expression by influencing transcript stability, splicing, translation efficiency and cap-independent translation, and in promoting circular RNA translation [review: (7)]. Mutations in many human genes encoding RNA modification enzymes have been linked to diseases, such as cancer, cardiovascular diseases, metabolic diseases, neurological disorders, and mitochondria-related defects [review: (8)].
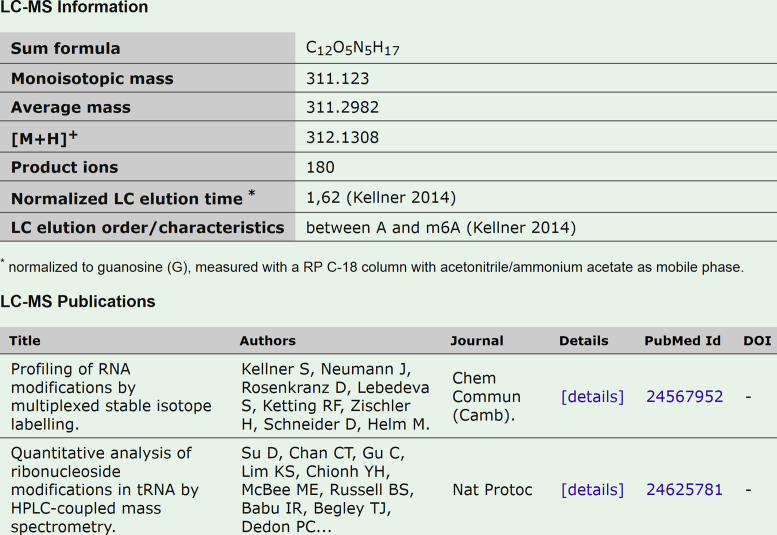
To adequately represent the recent accumulation of knowledge, we have added both to the variety and volume of data in the MODOMICS database. The most significant additions are: (i) extensively updated datasets: new modifications, new enzymes, and new RNA sequences with modifications; (ii) a new category of LC/MS data for modifications (Figure 1); (iii) new naming/numbering convention for modified residues in RNA sequences; (iv) replacement of Jmol by JSmol for 3D structure viewing.
Figure 1.
Example of MS/LC data for N2,N2-dimethylguanosine (m2,2G). The display of a new data type: LC/MS information on the modification detail page.
DATABASE CONTENT
MODOMICS has been developed to house and distribute collections of RNA modification pathways, chemical structures of modified nucleosides, sequences of modified RNAs, enzymes responsible for individual reactions, a catalog of ‘building blocks’ for chemical synthesis of modified RNA, and to be expanded to include new data types. The database was created as a single resource to organize and present all these data in a convenient and straightforward way and is currently the most comprehensive source of information among all existing RNA modification databases. Information about modified residues is also available in the RNAMDB database (9), while information about modified nucleosides identified from high-throughput experiments like Pseudo-seq, CeU-seq, m6A-seq, Aza-IP and RiboMeth-seq is hosted by RMBase (10). Recently MODOMICS was linked to RNAcentral, a database of non-coding RNA sequences (11), and serves as a source of modified tRNA and rRNA sequences.
At present, MODOMICS contains 163 different modifications that have been identified in RNA molecules. A typical entry for a modified ribonucleoside contains information about its fundamental chemical properties, chemical structure, localization in known RNA molecule types, the phylogenetic distribution with respect to Domains of Life, and known enzymes responsible for its biosynthesis. Among other available details information related to MS analyses of modified RNAs is provided (see the ‘LC/MS data for modified nucleosides’ section). Many of the products of modification reactions are substrates for further reactions, and the formation of hypermodified residues occurs in complex pathways, which are displayed as graphs in the PATHWAYS section of the database. Pathways are divided into six different categories according to their starting point: four categories correspond to the standard bases (A, G, C and U), one presents the incorporation and hypermodification pathway of queuosine, and one the modifications of the RNA 5′ cap.
MODOMICS provides a collection of modified RNA sequences of different types. For families of homologous RNAs, multiple sequence alignments are available. Sequences are visualized with all modifications highlighted and linked to the corresponding modification records. The current set of sequences comprises 691 tRNA, 19 rRNA, 46 snRNA and 25 snoRNA sequences.
The MODOMICS database currently contains information about 340 functionally characterized proteins involved in RNA modification, both functional enzymes and protein co-factors necessary for multi-protein enzymatic activities. For each protein a set of detailed information is provided and includes: identifiers and accession numbers from relevant resources and databases such as: NCBI GI, UniProt ID, COG number, PDB ID of structure (if available); amino acid sequence; corresponding ORF; information about catalyzed reaction, the position of modification and modified RNA(s) (if available). For proteins that are parts of enzymatic complexes, the name of the complex is provided.
MODOMICS also contains human and yeast snoRNAs, involved in RNA-guided RNA modification by the C/D box and H/ACA box ribonucleoproteins, linked to the corresponding modification sites in human and yeast RNAs and the catalog of ‘building blocks’ for the chemical synthesis of naturally occurring modified nucleosides.
Several options for database searching and querying are implemented in MODOMICS, including the BLAST (12) search of protein sequences and the PARALIGN (13) search of nucleic acid sequences collected in MODOMICS, as well as a utility that sends a protein sequence from a MODOMICS entry to BLAST on the NCBI web server.
Updated modifications section
Since the previous release of MODOMICS (14), 19 new modifications were added to the database. Among those are four types of geranylated nucleosides discovered in bacterial tRNA (3), 5-cyanomethyluridine (cnm5U) (15), and 2′-O-methyluridine 5-oxyacetic acid methyl ester (mcmo5Um) (5). LC/MS analyses of tRNAs from Bacillus subtilis, plants, and Trypanosoma brucei revealed the presence of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A), a derivative of N6-threonylcarbamoyladenosine (t6A), at position 37 of tRNAs responsible for recognition of adenosine-starting codons (16).
3D chemical structures of modified nucleosides are now displayed with JSmol (17). It is an open-source JS library and HTML5 viewer for 3D chemical structures which, in contrast to the previously used Jmol tool, does not require the installation of the Java software package. JSmol can also be used on systems that no longer support Java applets due to security concerns or for which Java is not available, like smartphones or tablets, and it does not use hardware graphics acceleration, enabling the software to run in any browser that supports HTML5 standards.
LC/MS data for modified nucleosides
This new MODOMICS release features a new section on modification detail page created to host the LC/MS data of the modified nucleoside. The new fields include information concerning the product ions, the protonated mass [M+H]+, the LC elution order and its characteristics, the normalized LC elution time and their literature references. The LC elution time is normalized to guanosine (G), measured with an RP C-18 column with acetonitrile/ammonium acetate as mobile phase and the elution order is based on the retention times of C, U, G, A and m6A to cover all areas of the chromatogram. The LC data is intended to provide the novice LC-MS user with guidance on the relative hydrophobicity of modified nucleosides and an estimated elution region using the denoted stationary and mobile phases. Currently, only 48 modifications, for instance, m1A, m3C, and m2,2G are associated with LC information, while 138 modifications have been associated with MS information. The new system is in place to be extended to all the modifications present in the database as soon as the new data become available. LC/MS based methods allow RNA modification profiling of different organisms in a semi-quantitative manner for the newly-detected modifications along with known modifications, and one can expect that approach will be further extended. LC/MS data for MODOMICS collection of modified nucleosides provides a comprehensive source of information for mapping of the identity and position of modified residues in RNA sequences.
New nomenclature for modified nucleosides
The old systems used to encode modified residues in RNA sequences have been very cumbersome for automated data processing, especially in the case of special characters (often interpreted as special actions) or names that contained letters such as ‘c’ or ‘i’ that could be confused with different bases. Thus, we developed a new naming convention that uses only digits in addition to standard letters (A, G, C, U), and which makes names distinct from one another. In the new proposed system, a number is introduced before the modified residue, so the software for sequence processing can recognize the original residue type before modification, as well as identify the specific modification(s) introduced. For the most common modification type, i.e., simple methylations, we use single digit numbers that indicate, whenever possible, methylated positions, with 0 representing the 2′-OH group. Consequently, modifications Am, m1A, m6A, m5C, are indicated as 0A, 1A, 6A, 5C, respectively. Residues with several methylations list all methylated positions, sorted in ascending order, e.g., m1Am, m2,2,7G, m4,4Cm are indicated as 01A, 227G, 044C, respectively. Some other modifications also use single digits for convenience, e.g., I and Ψ are indicated as 9A and 9U. Other modifications are indicated with additional numbers, usually following the position of the modification. For example, i6A, io6A, k2C, are indicated as 61A, 60A, 21C, respectively. Some naming decisions, especially in case of very complex modifications, were arbitrary, keeping in mind that ambiguity in the numbering must always be avoided, i.e., that a given sequence of digits preceding a letter corresponds to a unique modification. For each modified nucleoside the code in new nomenclature is available on the nucleoside site in the Modifications section of the database. The nomenclature was also implemented in the Sequences section by providing the option to display modifications in sequences using this nomenclature instead of one-letter symbols. As the next step we intend to develop and provide format conversion tools to allow for exporting and importing RNA sequences with modifications, and e.g., to run sequence searches that take modifications into account.
tRNA sequences section update and development
For this database release, 102 new tRNA sequences were added, and a major update of mammalian tRNA modifications was performed based on (18). Among new sequences are sets of tRNAs from two bacterial species: Streptomyces griseus and Lactococcus lactis (19,20), 60 and 26 sequences, respectively. As technology and methods improve, well-studied sequences continue to undergo revision (21). We have also introduced links of human tRNA sequences to MINTbase (22), which is a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. The MINTbase link in MODOMICS opens a page with a list of the latest profile of expressed tRNA fragments, aligned against the sequence from MODOMICS.
Updated collection of proteins, enzymatic activities, and pathways
The MODOMICS collection of functionally characterized proteins involved in RNA modification is under constant development. Since the previous release, 66 new proteins have been added. The collections of protein sequences and enzymatic activities are updated in parallel, which resulted in 105 new enzymatic activities. Among new proteins that were added in this release there is a collection of human RNA modification enzymes, including: dimethyladenosine transferase TFB1M (23), tRNA pseudouridine synthase PUS3 (24), tRNA m5C methyltransferases NSUN3 (25) and NSUN6 (26), and rRNA m5C methyltransferase NSUN4 (27). Apart from the addition of newly characterized enzymes, data entries for many enzymes and associated pathways were updated.
Future prospects
The number of experimentally identified modifications and RNA modifying enzymes keeps growing. New modified nucleosides are being discovered in particular in RNAs from recently adopted model systems, such as extremophilic prokaryotes. Though there is a considerable amount of information available about the enzymes responsible for introducing specific modifications, there are still many modified positions in well-characterized RNA molecules, for which the responsible enzymes are not known, e.g., m6Am at the 5′ end of human mRNAs or m5U, m4C in 12S mitochondrial rRNA. To help us keep up with new discoveries, we encourage the users of MODOMICS to submit suggestions for additions to be included in the database. We also encourage developers of other computational resources to contact us to have our databases mutually linked to each other.
For the next release of MODOMICS, we plan to update the visualization options and to refurbish the website, to keep up with the changing trends in web design. We also intend to renew data structures, to make MODOMICS more compatible with other databases and web servers, to facilitate automated data exchange, and to introduce the ability to search sequences by taking modifications into account.
AVAILABILITY
The data are accessible freely for research purposes at http://modomics.genesilico.pl.
ACKNOWLEDGEMENTS
We are grateful to Henri Grosjean, the co-founder, key co-supervisor, and key co-developer of MODOMICS, for his contribution to all previous releases. We thank Sebastian Leidel and Peter Dedon for discussions and useful suggestions. We also thank all other previous contributors to MODOMICS, for their work which provided a solid basis for the present update. We thank all members of the Bujnicki laboratory for fruitful discussions and useful suggestions. We are indebted to the authors of primary databases and services, whose content could be reused or linked to by MODOMICS. Last, but not least, we thank all users of MODOMICS who provided feedback and made suggestions, and who cited MODOMICS in their publications.
FUNDING
Polish National Science Center [NCN, 2012/04/A/NZ2/00455 to J.M.B, 2011/03/D/NZ1/03247 to E.P.]; IIMCB [statutory funds to J.M.B.]; National Institutes of Health [NIH, GM70641 to V. dC.-L, GM058843 to P.A.L.]; COST action [CA16120 ‘EPITRAN’ to J.M.B. and M.H.]; Deutsche Forschungsgemeinschaft [DFG, SPP1784/HE3397/13-1 to M.H.]. Funding for open access charge: Polish National Science Center [2012/04/A/NZ2/00455 to J.M.B.].
Conflict of interest statement. Janusz M. Bujnicki is an Executive Editor of Nucleic Acids Research.
REFERENCES
- 1. Grosjean H. Fine-tuning of RNA Functions by Modification and Editing. 2005; Berlin-Heidelberg: Springer-Verlag. [Google Scholar]
- 2. Lewis C.J., Pan T., Kalsotra A.. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol Cell Biol. 2017; 18:202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dumelin C.E., Chen Y., Leconte A.M., Chen Y.G., Liu D.R.. Discovery and biological characterization of geranylated RNA in bacteria. Nat. Chem. Biol. 2012; 8:913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zorbas C., Nicolas E., Wacheul L., Huvelle E., Heurgue-Hamard V., Lafontaine D.L.. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell. 2015; 26:2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sakai Y., Miyauchi K., Kimura S., Suzuki T.. Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res. 2016; 44:509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan C.T., Dyavaiah M., DeMott M.S., Taghizadeh K., Dedon P.C., Begley T.J.. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010; 6:e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roignant J.Y., Soller M.. m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017; 33:380–390. [DOI] [PubMed] [Google Scholar]
- 8. Jonkhout N., Tran J., Smith M.A., Schonrock N., Mattick J.S., Novoa E.M.. The RNA modification landscape in human disease. RNA. 2017; doi:10.1261/rna.063503.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cantara W.A., Crain P.F., Rozenski J., McCloskey J.A., Harris K.A., Zhang X., Vendeix F.A., Fabris D., Agris P.F.. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011; 39:D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun W.J., Li J.H., Liu S., Wu J., Zhou H., Qu L.H., Yang J.H.. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016; 44:D259–D265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Petrov A.I., Kay S.J., Gibson R., Kulesha E., Staines D., Bruford E.A., Wright M.W., Burge S., Finn R.D., Kersey P.J. et al. RNAcentral: an international database of ncRNA sequences. Nucleic Acids Res. 2015; 43:D123–D129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rognes T. ParAlign: a parallel sequence alignment algorithm for rapid and sensitive database searches. Nucleic Acids Res. 2001; 29:1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mandal D., Kohrer C., Su D., Babu I.R., Chan C.T., Liu Y., Soll D., Blum P., Kuwahara M., Dedon P.C. et al. Identification and codon reading properties of 5-cyanomethyl uridine, a new modified nucleoside found in the anticodon wobble position of mutant haloarchaeal isoleucine tRNAs. RNA. 2014; 20:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang B.I., Miyauchi K., Matuszewski M., D’Almeida G.S., Rubio M.A.T., Alfonzo J.D., Inoue K., Sakaguchi Y., Suzuki T., Sochacka E.. Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs. Nucleic Acids Res. 2017; 45:2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L.. JSmol and the next-generation web-based representation of 3D molecular structure as applied to proteopedia. Israel J Chem. 2013; 53:207–216. [Google Scholar]
- 18. Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014; 42:7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puri P., Wetzel C., Saffert P., Gaston K.W., Russell S.P., Cordero Varela J.A., van der Vlies P., Zhang G., Limbach P.A., Ignatova Z. et al. Systematic identification of tRNAome and its dynamics in Lactococcus lactis. Mol. Microbiol. 2014; 93:944–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cao X., Limbach P.A.. Enhanced detection of post-transcriptional modifications using a mass-exclusion list strategy for RNA modification mapping by LC-MS/MS. Anal. Chem. 2015; 87:8433–8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Addepalli B., Limbach P.A.. Pseudouridine in the anticodon of Escherichia coli tRNATyr(QPsiA) is catalyzed by the dual specificity enzyme RluF. J. Biol. Chem. 2016; 291:22327–22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pliatsika V., Loher P., Telonis A.G., Rigoutsos I.. MINTbase: a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics. 2016; 32:2481–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Metodiev M.D., Lesko N., Park C.B., Camara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C.M., Larsson N.G.. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009; 9:386–397. [DOI] [PubMed] [Google Scholar]
- 24. Shaheen R., Han L., Faqeih E., Ewida N., Alobeid E., Phizicky E.M., Alkuraya F.S.. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 2016; 135:707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van Haute L., Dietmann S., Kremer L., Hussain S., Pearce S.F., Powell C.A., Rorbach J., Lantaff R., Blanco S., Sauer S. et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 2016; 7:12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haag S., Warda A.S., Kretschmer J., Gunnigmann M.A., Hobartner C., Bohnsack M.T.. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA. 2015; 21:1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Metodiev M.D., Spahr H., Loguercio Polosa P., Meharg C., Becker C., Altmueller J., Habermann B., Larsson N.G., Ruzzenente B.. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014; 10:e1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]