Abstract
TADB2.0 (http://bioinfo-mml.sjtu.edu.cn/TADB2/) is an updated database that provides comprehensive information about bacterial type II toxin–antitoxin (TA) loci. Compared with the previous version, the database refined and the new data schema is employed. With the aid of text mining and manual curation, it recorded 6193 type II TA loci in 870 replicons of bacteria and archaea, including 105 experimentally validated TA loci. In addition, the newly developed tool TAfinder combines the homolog searches and the operon structure detection, allowing the prediction for type II TA pairs in bacterial genome sequences. It also helps to investigate the genomic context of predicted TA loci for putative virulence factors, antimicrobial resistance determinants and mobile genetic elements via alignments to the specific public databases. Additionally, the module TAfinder-Compare allows comparing the presence of the given TA loci across the close relative genomes. With the recent updates, TADB2.0 might provide better support for understanding the important roles of type II TA systems in the prokaryotic life activities.
INTRODUCTION
Toxin–antitoxin (TA) systems, initially identified as plasmid addiction modules, are highly abundant on the chromosomes of most free-living bacteria (1). The TA systems are involved in multiple life activities of bacteria, such as nutrition starvation (1,2), programmed cell death (3), protection from bacteriophage (4) and the antimicrobial resistance (5). TA system consists of a stable toxin protein and a labile cognate antitoxin encoded by a bicistronic locus. Depending on the molecular pattern of antitoxin and the mechanism of toxin neutralization, the known TA systems have been sorted into six different groups, namely type I to VI (6,7). The antitoxin molecules are small non-coding RNAs in type I and III TA pairs, while they are proteins in other types. Toxins act on different targets to affect various cellular processes (Supplementary Table S1), such as peptidoglycan synthesis, replication and translation. Recently, two type II toxins containing the Gcn5-related N-acetyltransferase (GNAT) domain, TacT of Salmonella enterica Typhimurium (8) and AtaT of Escherichia coli O157:H7 (9), were reported to transfer the acetyl group from acetyl coenzyme A to the amine group of the tRNAs, resulting in halting translation of bacterial cell. Thus, more diverse mechanisms of how toxin works and gets neutralized by antitoxin are pending to be elucidated (10).
Among all the six types of TA system, type II is the most extensively studied, with large quantity and high quality of publicly accessible data. With the advent of next-generation sequencing, a vast number of bacterial genome sequences are being generated. The knowledge resource systematically collecting the reported type II TA loci is therefore needed for researchers to sharing data, gaining reference and predicting new TA pairs. There are currently two open-access bioinformatics resources in the field of type II TA loci, namely the online tool RASTA (11) and the web-based database TADB (12). RASTA searches the conserved functional domains of individual toxins or antitoxin proteins; however, the RASTA website announced that its maintenance stopped since 2011 (http://genoweb1.irisa.fr/duals/RASTA-Bacteria/). In the year of 2011, we proposed the web-based open-access database TADB1.1 archiving both the experimentally validated type II TA pairs and the data derived from computationally predicted datasets (13,14). Since then, a vast number of new TA pairs were identified experimentally. The demand for database update and a tool to help predict new TA pairs became urgent.
Here, we report the new major release of TADB, version 2.0. It reflects a large increase in the curated dataset of known type II TA pair and reorganization of the data schema. A newly developed online prediction tool for type II TA loci was also integrated. We expect that TADB2.0 will provide a better support for researchers interested in bacterial type II TA systems.
MATERIALS AND METHODS
Data update by text mining and manual curations
The maintenance of TADB core dataset has been performed majorly on addition and proof of the TA loci experimental evidence. The TA pair was tagged as ‘experimentally validated’ in TADB2.0 only if its clear biological function was reported in a peer-reviewed scientific publication. After manual curation of the PubMed searching results with the key word of ‘toxin and antitoxin’, 326 papers published since 2011 were collected and added into TADB2.0, resulting in a total of 586 papers in the database. TADB2.0 built the links of the archived type II TA loci to the corresponding experimental literature. For the TA loci with experimental evidence, in the previous version, the mapping was set between the archived TA loci and the literature on the host strain level. Now, the link was straightforwardly built from the TA loci to the corresponding experimental literature. In TADB2.0, 105 pairs of experimentally validated type II TA loci were collected.
In addition, TADB2.0 also recorded 6088 computationally predicted type II TA loci. They were taken from two datasets: set I containing the BLASTp-identified TA pairs reported by Pandey et al. (13); Set II including the TA loci assigned to 44 conserved TA domain pairs proposed by Makarova et al. (14). However, note that, contrary to the previous version, TADB2.0 had not recorded the third in silico dataset obtained from RASTA-Bacteria (11). Namely, RASTA-Bacteria employed RPSBLAST searches to detected the individual toxin or antitoxin proteins; however, the obtained toxin or antitoxin genes were paired by TADB1.1 without the proven accuracy (12). The updated version had thus removed the RASTA-Bacteria dataset and just showed them as the supplemental list on the web pages of browsing individual replicons. The interface of TADB2.0 data presentation is illustrated in Supplementary Figure S1.
New schema of the back-end database
Similarly, to the previous version, TADB2.0 is implemented as a PostgreSQL relational database, but the database tables were constructed with a new data schema. In TADB1.1, the Generic Model Organism Database’s Chado schema (15) was employed to house the data of ∼1000 prokaryotic genomes obtained from the NCBI RefSeq project archive, sequence and annotation included. As the available genome data blasted in the recent years, TADB1.1 turned out to be hard to maintain and update. In the present release, a new local schema of PostgreSQL was designed to reorganize the TA locus data efficiently. It contains two major modules: TA pair (Supplementary Figure S2) and external reference. By this, the number of tables in the back-end of our database has been decreased to 21, compared to 194 in the previous version. The view number also decreased from 83 to 4. The file storage space thus reduced from 26 Gb to ∼31 Mb.
Integration of the type II TA loci prediction tool
Conserved genetic organization of known type II TA systems typically contains two tandem genes coding for cognate protein partners. We integrated a newly developed online tool, named TAfinder, into TADB2.0 to quickly detect the putative type II TA loci in bacterial genome sequences. In the previous release, the prediction of the putative toxin or antitoxin loci was aided by homolog searches by using the TADB-host WU-BLAST tool or the RPSBLAST-based tool RASTA. In TADB2.0, the TAfinder combines the homolog search module and the operon structure detection module, allowing the enhancement of prediction performance for type II TA pair (Supplementary Figures S3 and S4). Briefly, the TAfinder starts at searching the toxin or antitoxin protein homologs by using both NCBI BLASTp (16) and HMMer3 (17). The BLASTp subject dataset contains all the TADB2.0-archived type II TA systems. The 108 HMM-profiles for the conserved toxin domains and 201 for the antitoxin domains are also detected by HMMer3 with the default settings. Then, the short homologs of toxin proteins or antitoxin proteins with the length of 30–200 amino acid residues were kept as candidates. Finally, two flanking toxin and antitoxin candidate genes on the same DNA strand and with the intergenic distances of −20 to 150 bp were paired as an operon structure, thus predicted as a putative Type II TA locus. The Perl/Bioperl scripts have been written to parse HMMer/BLASTp search results and co-localize significant hits efficiently on a local Linux server.
The TAfinder tool was designed to contain two functional modules, TAfinder-Predict and TAfinder-Compare. For the TAfinder-Predict module, the input data in multiple types are acceptable, including the amino acid sequence, nucleotide sequence, the annotated GenBank file and even the unannotated sequences of scaffolds or contigs. For input file with nucleotide sequence, the TAfinder users are encouraged to submit the annotated data, preferably with manual curation, for the maximum accuracy. But the raw nucleotide sequence could be also accepted. It would be preprocessed by our quick gene annotation tool CDSeasy (18), and then used as an input to TAfinder for TA prediction. We have also downloaded the genomic data of 5184 completely sequenced replicons. Users could select the data of interest either by ticking in the genome list or if applicable, typing in the RefSeq accession number. The output interface of the TAfinder-Predict module presents the putative TA pairs in tabular form on the web, and provides the hyperlink to other databases publicly available, such as NCBI, to meet the users’ demands. The TAfinder-Predict module also helps to investigate the genomic context of predicted TA loci for virulence factor, antimicrobial resistance determinants and genes related to horizontal transfer via alignments to the specific public databases (Supplementary Figure S3). Additionally, the TAfinder-Compare module allows doing the alignment between the identified TA loci in multiple closely related bacterial genomes. Finally, TAfinder-Compare measures the toxin or antitoxin protein sequence similarities by using BLASTp-based H-value (19,20) (Supplementary Figure S5).
RESULTS AND DISCUSSION
The web-based database TADB1.1 published in 2011 (12) has been offering a comprehensive compilation of both in silico predicted and experimentally supported type II TA locus data and genetic features. As a unique TA database, it has been cited more than 100 times. Recently a number of new type II TA pairs and other type TA systems have been characterized experimentally, suggesting an urgent demand for the updated version. Recent developments present in this study have further improved the data quality of TADB, such as the type II TA loci curation with experimental literature support. Compared to TADB1.1, the updated TADB2.0 offers three major improvements: (i) new type II TA loci dataset via manual curation; (ii) new data schema of the back-end database; (iii) online tool for type II TA pair prediction.
Experimentally validated dataset provided by TADB could be applicable to detect putative type II TA loci in a wide range of bacterial species. For example, there is no well documented relBE family of type II TA systems reported in Streptomyces. We thus searched for the relBE locus against eight completely sequenced Streptomyces genomes based on the conserved RelBE domains, and then identified and characterized a new chromosomal relBE locus in Streptomyces cattleya DSM46488 (21). We also examined ten completely sequenced Klebsiella pneumoniae genomes and 212 putative type II TA loci were identified, including 77 toxin proteins containing GNAT conserved domain (22). In this study, extending on our local Perl/Bioperl scripts, we developed user-friendly tool TAfinder as a public resource for detecting type II TA pairs in bacterial genome sequences. It employed the training dataset taken from TADB2.0. With respect to RASTA based on individual toxin or antitoxin domain searches, TAfinder was designed to identify the TA pair to facilitate functional interpretation. It combines homolog searches and operon feature detection (Supplementary Figure S5). To date, TAfinder has been applied in silico analysis and/or the support of the experimental validations in several bacteria by other research groups, including Staphylococcus aureus (23), Acinetobacter baumannii (24) and the IncX plasmids (25). While the TADB2.0 browse module is still functioning (to accommodate the experimentally validated TA loci), for predicting putative type II TA loci, we strongly encourage users to run TAfinder instead of only browsing the TADB2.0 web page.
Comprehensive genome in silico analysis revealed that the type II TA systems are diverse and widespread in the prokaryotic organisms (26). After the examination of 2786 species of prokaryotes with the publicly available complete genome sequences (Supplementary Figure S6 and Table S2), TAfinder prediction results showed that 66% of species harbored 1–20 type II TA loci in individual strains while 20% of species carried more than 20 loci. The cross-talks of the intra- and inter-type TA systems can be explored (23). Remarkably, there are no type II TA loci detected in 14% of species, including Prochlorococcus marinus and the small Mycoplasma symbionts. Whether the TA systems might promote the bacterial fitness and facilitate the evolution of free-living organisms awaits investigation within the population. In addition, among all type II TA loci of the organisms under study, the top five conserved domains of the toxin proteins were RelE, PIN, MNT, MazF and VapC (Supplementary Figure S7A). But the distribution biases might change for the different species; for example, the number of the GNAT toxins in S. enterica (Supplementary Figure S7B) and K. pneumoniae (Supplementary Figure S7C) rises dramatically, just following that of the RelE toxins. In their corresponding antitoxins, the DNA binding-associated domains HTH, RHH and DUF1778 were usually found.
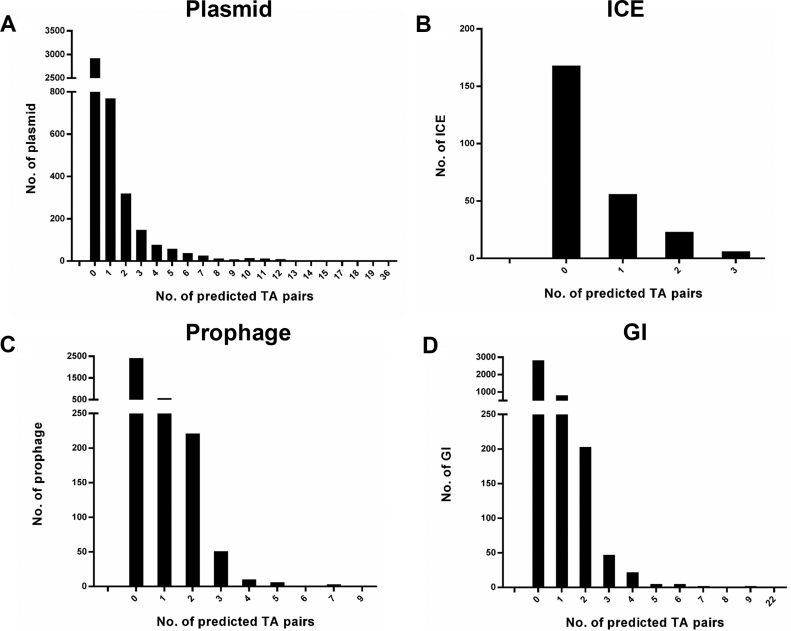
A few plasmid-encoded TA systems have been well documented, providing competitive advantage to the host. Out of the 4419 (2165 from whole genome sequence data +2254 from independent plasmid sequencing) plasmids with the whole sequences (Supplementary Table S2), 33.9% (1499/4419) plasmids carried 3592 TAfinder-predicted type II TA loci. Additionally, the TA loci have also been increasingly discovered within or close to the mobile elements on bacterial chromosomes, including prophages, genomic islands (GIs) and integrative and conjugative elements (ICEs) (27,28). Via the TAfinder predictions with the input from multiple data sources (Supplementary Table S2), 33.6% (85/253) ICEs had 120 putative type II TA loci, 26.3% (863/3278) prophages carried 1237 type II TA loci and 28.1% (1099/3918) GIs host 1563 type II TA loci (Figure 1 and Supplementary Figures S8). These type II TA systems might play key roles in the stabilization of the self-transmissible integrative elements, which thus contribute to the horizontal transfer of virulence factors, antibiotic-resistant determinants and many other important bacterial traits.
Figure 1.
The number of TAfinder-predicted type II TA loci in the mobile genetic elements. Panel (A), (B), (C) and (D) shows the type II TA number for 4419 plasmids, 253 integrative conjugative elements (ICEs), 3278 prophages and 3918 genomic islands (GIs) under analysis (Supplementary Table S2), respectively.
CONCLUSION
Here, we reported a major upgrade of TADB, where the type II TA loci have been collected in TADB2.0, with an optimized database back-end organization. The unique type II TA system prediction tool has been integrated into this database, aiding researcher to explore new TA pairs in the high-speed growing bacterial genomic data. New records of publications will be mined to update the status of currently archived TA systems. Newly available data on TA loci will be uploaded regularly to keep pace with the rapidly expanding bacterial genome database. Finally, we propose an updated type II TA-specific resource which is expected to facilitate efficient investigation of large numbers of these systems, recognition of patterns corresponding to cellular targets of diverse toxins, and the neutralization mechanism of the cognate antitoxin, and an improved understanding of their biological roles and significance.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31371261, 31670074]; Ministry of Science and Technology, China (973 program) [2015CB554202]. Funding for open access charge: Ministry of Science and Technology, China (973 program) [2015CB554202].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gerdes K., Maisonneuve E.. Bacterial persistence and toxin–antitoxin loci. Annu. Rev. Microbiol. 2012; 66:103–123. [DOI] [PubMed] [Google Scholar]
- 2. Harms A., Maisonneuve E., Gerdes K.. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016; 354:aaf4268–aaf4269. [DOI] [PubMed] [Google Scholar]
- 3. Hu M.-X., Zhang X., Li E.-L., Feng Y.-J.. Recent advancements in toxin and antitoxin systems involved in bacterial programmed cell death. Int. J. Microbiol. 2010; 2010:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fineran P.C., Blower T.R., Foulds I.J., Humphreys D.P., Lilley K.S., Salmond G.P.C.. The phage abortive infection system, ToxIN, functions as a protein–RNA toxin–antitoxin pair. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang Q.E., Walsh T.R.. Toxin–antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol. Rev. 2017; 41:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lobato-Márquez D., Díaz-Orejas R., García-Del Portillo F.. Toxin–antitoxins and bacterial virulence. FEMS Microbiol. Rev. 2016; 40:592–609. [DOI] [PubMed] [Google Scholar]
- 7. Page R., Peti W.. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016; 12:208–214. [DOI] [PubMed] [Google Scholar]
- 8. Cheverton A.M., Gollan B., Przydacz M., Wong C.T., Mylona A., Hare S.A., Helaine S.. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell. 2016; 63:86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jurėnas D., Chatterjee S., Konijnenberg A., Sobott F., Droogmans L., Garcia-Pino A., Van Melderen L.. AtaT blocks translation initiation by N-acetylation of the initiator tRNAfMet. Nat. Chem. Biol. 2017; 13:640–646. [DOI] [PubMed] [Google Scholar]
- 10. Hall A.M., Gollan B., Helaine S.. Toxin-antitoxin systems: reversible toxicity. Curr. Opin. Microbiol. 2017; 36:102–110. [DOI] [PubMed] [Google Scholar]
- 11. Sevin E.W., Barloy-Hubler F.. RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007; 8:R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shao Y., Harrison E.M., Bi D., Tai C., He X., Ou H.Y., Rajakumar K., Deng Z.. TADB: A web-based resource for type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011; 39:606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pandey D.P. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005; 33:966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Makarova K.S., Wolf Y.I., Koonin E. V. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009; 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mungall C.J., Emmert D.B.. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007; 23:i337–i346. [DOI] [PubMed] [Google Scholar]
- 16. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R.. HMMER web server: 2015 update. Nucleic Acids Res. 2015; 43:W30–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J., Tai C., Deng Z., Zhong W., He Y., Ou H.-Y.. VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief. Bioinform. 2017; doi:10.1093/bib/bbw141. [DOI] [PubMed] [Google Scholar]
- 19. Fukiya S., Mizoguchi H., Tobe T., Mori H.. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 2004; 186:3911–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shao Y., He X., Harrison E.M., Tai C., Ou H.-Y., Rajakumar K., Deng Z.. mGenomeSubtractor: a web-based tool for parallel in silico subtractive hybridization analysis of multiple bacterial genomes. Nucleic Acids Res. 2010; 38:W194–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li P., Tai C., Deng Z., Gan J., Oggioni M.R., Ou H.-Y.. Identification and characterization of chromosomal relBE toxin-antitoxin locus in Streptomyces cattleya DSM46488. Sci. Rep. 2016; 6:32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wei Y.Q., Bi D.X., Wei D.Q., Ou H.Y.. Prediction of type II toxin-antitoxin loci in Klebsiella pneumoniae genome sequences. Interdiscip. Sci. Comput. Life Sci. 2016; 8:143–149. [DOI] [PubMed] [Google Scholar]
- 23. Conlon B.P., Rowe S.E., Gandt A.B., Nuxoll A.S., Donegan N.P., Zalis E.A., Clair G., Adkins J.N., Cheung A.L., Lewis K.. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 2016; 1:16051. [DOI] [PubMed] [Google Scholar]
- 24. Michiels J.E., Van den Bergh B., Fauvart M., Michiels J.. Draft genome sequence of Acinetobacter baumannii strain NCTC 13423, a multidrug-resistant clinical isolate. Stand. Genomic Sci. 2016; 11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bustamante P., Iredell J.R.. Carriage of type II toxin-antitoxin systems by the growing group of IncX plasmids. Plasmid. 2017; 91:19–27. [DOI] [PubMed] [Google Scholar]
- 26. Leplae R., Geeraerts D., Hallez R., Guglielmini J., Drèze P., Van Melderen L.. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011; 39:5513–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wozniak R.A.F., Waldor M.K.. A toxin–antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009; 5:e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Melderen L., Saavedra De Bast M.. Bacterial toxin–antitoxin systems: more than selfish entities?. PLoS Genet. 2009; 5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.