Abstract
MoonProt 2.0 (http://moonlightingproteins.org) is an updated, comprehensive and open-access database storing expert-curated annotations for moonlighting proteins. Moonlighting proteins contain two or more physiologically relevant distinct functions performed by a single polypeptide chain. Here, we describe developments in the MoonProt website and database since our previous report in the Database Issue of Nucleic Acids Research. For this V 2.0 release, we expanded the number of proteins annotated to 370 and modified several dozen protein annotations with additional or updated information, including more links to protein structures in the Protein Data Bank, compared with the previous release. The new entries include more examples from humans and several model organisms, more proteins involved in disease, and proteins with different combinations of functions. The updated web interface includes a search function using BLAST to enable users to search the database for proteins that share amino acid sequence similarity with a protein of interest. The updated website also includes additional background information about moonlighting proteins and an expanded list of links to published articles about moonlighting proteins.
INTRODUCTION
MoonProt is an expert manually curated and non-redundant resource of information about moonlighting proteins. Moonlighting proteins are proteins in which more than one physiologically relevant discrete function is performed by a single polypeptide chain (1–3). For example, the taxon specific crystallins are lens structural proteins in the eyes of several species and a metabolic enzymes in other tissues (4). Moonlighting proteins are found throughout the evolutionary tree and perform many kinds of functions (1–11).
Moonlighting proteins are usually found through serendipity, lacking a shared sequence or structural feature that can indicate that a protein has multiple functions, and information about the proteins is scattered in many different publications, so a database provides a way for researchers to learn about these proteins and to find out if a protein of interest is a known moonlighting protein or related to a known moonlighting protein. In addition, the collection of information about known moonlighting proteins can aid in understanding the connections between protein structure and function, determining the functions of genes identified in newly sequenced genomes, interpreting proteomics results, and annotating protein sequence and structural databases. Information about the structures and functions of moonlighting proteins can be helpful in understanding the evolution of protein function, which can also help in the design of proteins with novel functions.
In 2014, our lab constructed the open-access web server MoonProt, the Moonlighting Proteins Database (http://www.moonlightingproteins.org/) (12). In this paper, we present the latest version of MoonProt. Since its first development three years ago, the database has grown to include annotation for 370 proteins, the website interface has been redesigned, and information about individual moonlighting proteins and moonlighting proteins in general have been updated.
MATERIALS AND METHODS
Selection of moonlighting proteins included in the database
For inclusion of a protein in the MoonProt Database, peer-reviewed published biochemical, biophysical, mutagenic, or other data to support the presence of multiple physiologically-relevant functions was required and was critically reviewed by the PI. Proteins were not included if the ‘multiple functions’ are due to gene fusions, different RNA splice variants, the same function in two different locations, pleiotropic effects on multiple pathways or multiple physiological processes, or a family of proteins in which the different functions are performed by different proteins. Proteins were not included if the ‘multiple functions’ are simply different aspects of the same function (i.e. ‘membrane protein’ and ‘transmembrane receptor’).
Information included about individual proteins
Information about each protein was manually curated from published journal articles and online resources as described for Version 1.0 (12). The entry for each protein includes a description of each function and a list of references for publications providing experimental evidence of that function. When available, information is included about the specific cellular location in which the protein exhibits each function. Importantly, the specific species in which each protein has two or more functions was identified and included because a homologue from another species might or might not have both functions. Amino acid sequences were identified using UniProtKB (13) or Pubmed [http://www.ncbi.nlm.nih.gov/pubmed/] resources and are included in FASTA format. Those sequences were used with BLAST [http://blast.ncbi.nlm.nih.gov/Blast.cgi] to identify structures in the Protein Data Bank (14) that correspond to the amino acid sequence, if available. GO terms (15) were identified from the UniProtKB (13), and Enzyme Commission (EC) numbers are included in order to illustrate the different types of proteins included. UniProt entry IDs are included as links for easy connection to external resources.
Database architecture and web interface
The database is based on MySQL (http://www.mysql.com) for data storage, together with PHP 7.1 (http://www.php.net), HTML (HyperText Markup Language), and CSS (Cascading Style Sheets) for construction of the new interface. A Content Management System (CMS): WordPress, which utilizes modern web technologies, was used to help streamline the software development process.
RESULTS
New developments in MoonProt
Additional proteins and updated annotations
The MoonProt Database version 2.0 is now available at www.moonlightingproteins.org and provides information about hundreds of moonlighting proteins for which experimental evidence is available confirming the presence of more than one function. The database has grown by over one third since our last report with an additional 90 moonlighting proteins added based on information from the peer-reviewed literature. At the time of writing, the database includes 370 proteins. The new entries increase the number of human proteins included to 73, with an increase in the number of proteins from several model organisms such as Saccharomyces cerevisiae (34 proteins) and Escherichia coli (31 proteins).
As in version 1.0, most of the new entries have catalytic activities as one or more of their functions. There is also an increase in the number of proteins that are enzymes or chaperones inside the cell and have a second function on the cell surface or when secreted to the extracellular fluid (i.e. blood). Many of these proteins play important roles in health and disease. For prokaryotes, cytoplasmic enzymes can have a second role as a secreted signaling protein that affects the host immune system or as a cell surface receptor for host proteins. This can play a key role in infection for pathogens, but even commensal or ‘good’ bacteria have been found to make use of intracellular/surface moonlighting proteins to interact with the host. Even our own cells make use of cytoplasmic proteins on the cell surface, such as in several new additions to the database that are cytosolic enzymes that are also found on the surface of sperm and involved in sperm and egg interactions during fertilization.
Along with adding more proteins to the database, the annotation for many of the proteins has been updated, including more links to protein structures in the Protein Data Bank. For some proteins, additional references have been included, and a few dozen outdated UniProt IDs have been replaced with updated IDs.
New web interface
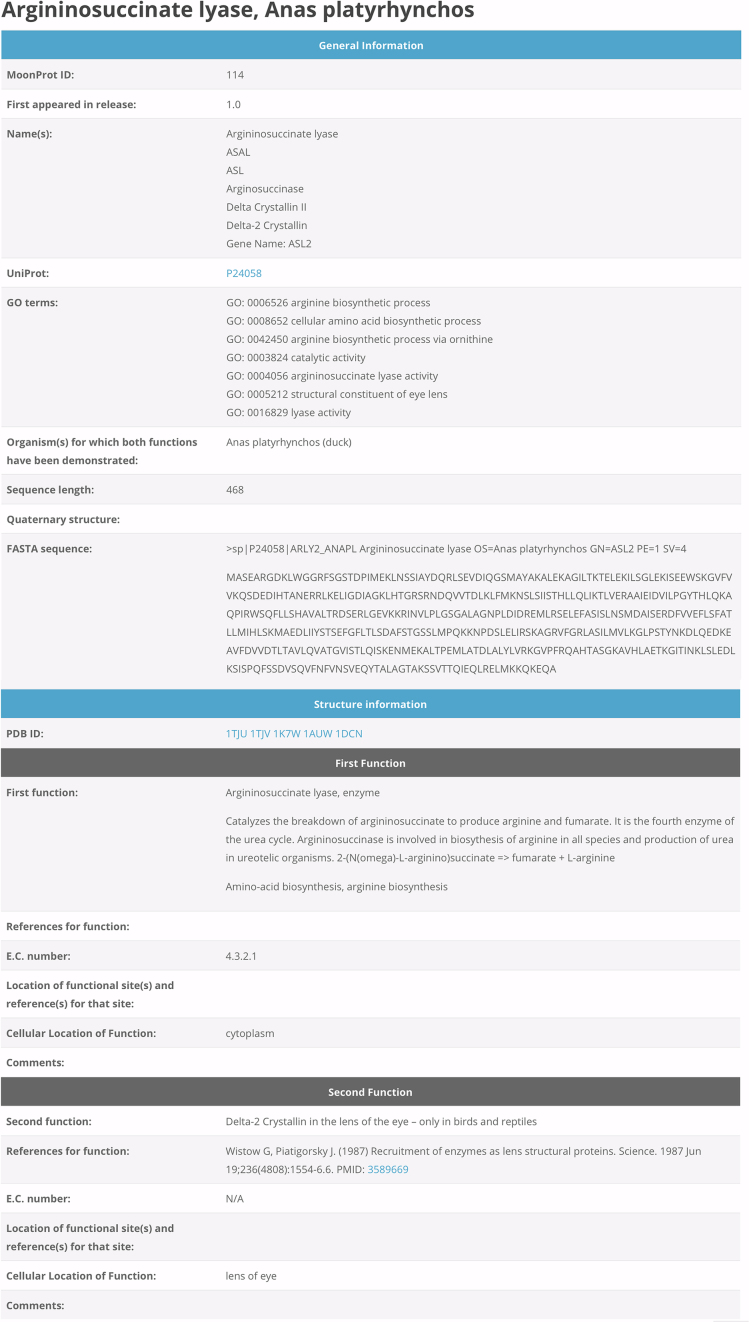
Since our last publication, we have developed a website with a new interface located at www.moonlightingproteins.org that gives access to the manually curated information about moonlighting proteins. The front page/home page, which is now also accessible with full functionality on mobile devices, includes a panel of summary information and several mechanisms to access the data. Several of the previous interaction options are also available, including a Proteins link that leads to a list of all the proteins in the database. Clicking on the protein names in the Proteins list will lead to the individual Protein Details page that displays the annotation information for that protein (Figure 1). Other links on the home page lead to general information about moonlighting proteins (FAQs), review articles about moonlighting proteins (Publications), and references for resources used in annotating the database (Resources). The information in each of these pages has been expanded and updated.
Figure 1.
Example of a protein annotation page. Each protein page contains the names of the protein, a UniProt accession number, the species of organism for which the protein has been shown to have more than one function (homologues of a moonlighting protein might have only one of the functions), GO terms, the length of the amino acid sequence, the amino acid sequence in FASTA format, PDB IDs for any available protein structures in the Protein Data Bank, descriptions of at least two functions, links to peer-reviewed publications describing experiments demonstrating the protein performs each function, and Enzyme Commission numbers (if an enzyme).
BLAST search function added
On the homepage, an updated Search link leads to a page with two types of search options, a text search and a BLAST sequence similarity search. The Search box enables a text search of all the annotated information in the database, which is expanded from the first version of the database, which allowed a search of only some of the categories of information. The search returns a list of protein entries containing that term.
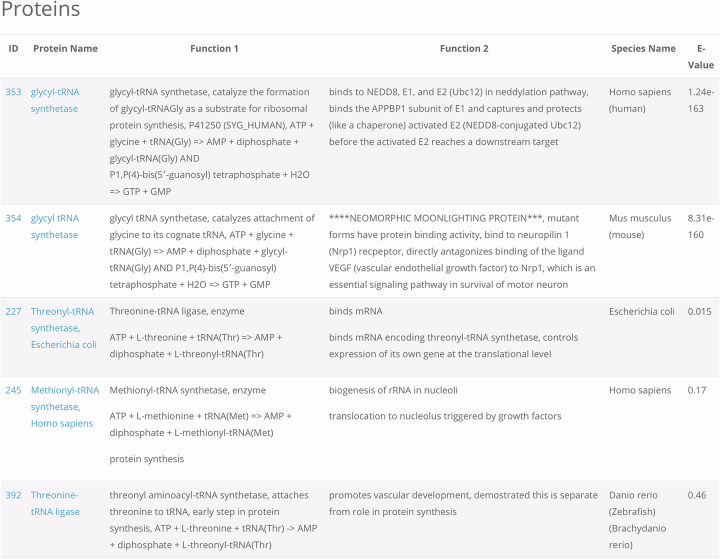
A second box on the Search page, labeled BLAST, enables use of the NCBI-blast-2.6.0+ algorithm (Basic Local Alignment Search Tool) (16) to search the database for moonlighting proteins that share sequence similarity with a query sequence. Users can paste an amino acid sequence (in the single letter code) in the box, and the search returns a sorted list of protein queries ranked by their similarity to the query sequence (Figure 2). By using this feature a user can determine if their protein of interest is a known moonlighting protein or if any of the known moonlighting proteins share sequence similarity to their protein of interest.
Figure 2.
Example of the output of a Blast query. Users can supply the amino acid sequence of a protein of interest and check if that protein or a homologous protein is in the MoonProt Database. In this example, the user submitted a fragment of the sequence for glycyl-tRNA synthetase, ‘FNLMFKTFIGPGGNMPGYLRPETAQGIFLNFKRLLEFNQGKLPFAAAQIGNSFRNEISPRSGLIRVREFTMAEIEHFVDPSEKDHPKFQNVADLHLYLYSAKAQVSGQSARKMRLGDAVEQGVINNTVLGYFIGRIYLYLTKVGISPDKLRFRQHMENEMAHYACDCWDAESKTSYGWIEIVGCADRSCYDLSCHARATKVPLVAEKPLKEPKTVNV’. The search returns a sorted list of protein names ranked by their similarity to glycyl-tRNA synthetase, the query sequence. Clicking on the link for each protein name leads to its protein page.
CONCLUSIONS AND PERSPECTIVES
The MoonProt Database version 2.0 is now available at www.moonlightingproteins.org and provides a centralized, organized resource containing information about 370 moonlighting proteins for which experimental evidence is available for more than one function.
Most moonlighting proteins have been discovered through serendipity, with the absence of a common physical or sequence characteristic among moonlighting proteins, which prevents the development of a robust algorithm for accurately predicting the presence of moonlighting functions. This database, with its collection of information about hundreds of moonlighting proteins, provides a resource for labs interested in developing computational methods for predicting protein functions based on sequence, structure, cellular localization, protein–protein interactions, or other characteristics. It also includes links to structures in the Protein Data Bank that could be used by synthetic biologists as a guide for designing proteins that can perform more than one function. We note that MoonProt 2.0 might be more useful for some of these purposes than another recent resource describing multifunctional proteins (17) because MoonProt only includes proteins for which biochemical or biophysical experiments demonstrated that the multiple functions are performed by a single polypeptide chain and are not due to different functions of different proteins within a large multiprotein complex or the effects of pleiotropy or other similar mechanisms.
We continue to add annotations to the MoonProt Database as new peer-reviewed publications about moonlighting proteins become available and as new protein structures are deposited in the Protein Data Bank. The MoonProt Database is likely to grow considerably in the next few years as the discovery of protein functions is aided by large scale functional proteomics studies. In addition, new formation about the known moonlighting proteins is likely to increase as new protein structures are solved.
AVAILABILITY AND LICENSE
The MoonProt Database is freely available via a user-friendly graphical user interface (GUI) at the web address www.moonlightingproteins.org. The interface enables text search for a protein name, species, or a UniProtKB or PDB identifier and a BLAST search using an amino acid sequence in the one letter code. The user can also browse a list of all the proteins in the database. The database is ‘read and search only’ by the public, but additional information about the known moonlighting proteins and suggestions of other proteins that might also be moonlighting are welcome and can be sent to the curators for possible inclusion in the database.
FUNDING
UIC College of Liberal Arts and Sciences Award for Faculty in the Natural Sciences (to C.J.J.). Funding for open access charge: UIC College of Liberal Arts and Sciences Award for Faculty in the Natural Sciences (to C.J.J.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Jeffery C.J. Moonlighting proteins. Trends Biochem. Sci. 1999; 24:8–11. [DOI] [PubMed] [Google Scholar]
- 2. Jeffery C.J. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 2003; 19:415–417. [DOI] [PubMed] [Google Scholar]
- 3. Jeffery C.J. Moonlighting proteins—an update. Mol. BioSystems. 2009; 5:345–350. [DOI] [PubMed] [Google Scholar]
- 4. Wistow G., Piatigorsky J.. Recruitment of enzymes as lens structural proteins. Science. 1987; 236:1554–1556. [DOI] [PubMed] [Google Scholar]
- 5. Guo M., Schimmel P.. Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 2013; 9:145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henderson B., Martin A.. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 2011; 79:3476–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henderson B., Pockley A.G.. Molecular chaperones and protein-folding catalysts as intercellular signaling regulators in immunity and inflammation. J. Leukoc. Biol. 2010; 88:445–462. [DOI] [PubMed] [Google Scholar]
- 8. Gancedo C., Flores C.L.. Moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 2008; 72:197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Commichau F.M., Stülke J.. Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol. Microbiol. 2008; 67:692–702. [DOI] [PubMed] [Google Scholar]
- 10. Piatigorsky J. Gene Sharing and Evolution. 2007; Cambridge: Harvard University Press. [Google Scholar]
- 11. Nobeli I., Favia A.D., Wool I.G.. Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 1996; 21:164–165. [PubMed] [Google Scholar]
- 12. Mani M., Chen C., Amblee V., Liu H., Mathur T., Zwicke G., Zabad S., Patel B., Thakkar J., Jeffery C.J.. MoonProt: a database for proteins that are known to moonlight. Nucleic Acids Res. 2014; 43:D277–D282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rose P.W., Prlić A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Constanzo L.D., Duarte J.M., Dutta S., Feng Z. et al. . The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2016; 45:D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gene Ontology Consortium Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017; 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McGinnis S., Madden T.L.. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004; 32:W20–W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hernandez S., Ferragut G., Amela I., Perez-Pons J.A., Pinol J., Mozo-Villarias A., Cedano J., Querol E.. MultitaskProtDB: a database of multitasking proteins. Nucleic Acids Res. 2014; 42:D517–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]