Abstract
The Human Metabolome Database or HMDB (www.hmdb.ca) is a web-enabled metabolomic database containing comprehensive information about human metabolites along with their biological roles, physiological concentrations, disease associations, chemical reactions, metabolic pathways, and reference spectra. First described in 2007, the HMDB is now considered the standard metabolomic resource for human metabolic studies. Over the past decade the HMDB has continued to grow and evolve in response to emerging needs for metabolomics researchers and continuing changes in web standards. This year's update, HMDB 4.0, represents the most significant upgrade to the database in its history. For instance, the number of fully annotated metabolites has increased by nearly threefold, the number of experimental spectra has grown by almost fourfold and the number of illustrated metabolic pathways has grown by a factor of almost 60. Significant improvements have also been made to the HMDB’s chemical taxonomy, chemical ontology, spectral viewing, and spectral/text searching tools. A great deal of brand new data has also been added to HMDB 4.0. This includes large quantities of predicted MS/MS and GC–MS reference spectral data as well as predicted (physiologically feasible) metabolite structures to facilitate novel metabolite identification. Additional information on metabolite-SNP interactions and the influence of drugs on metabolite levels (pharmacometabolomics) has also been added. Many other important improvements in the content, the interface, and the performance of the HMDB website have been made and these should greatly enhance its ease of use and its potential applications in nutrition, biochemistry, clinical chemistry, clinical genetics, medicine, and metabolomics science.
INTRODUCTION
The Human Metabolome Database (HMDB) is a comprehensive, freely available web resource containing detailed information about the human metabolome. The human metabolome represents the complete collection of small molecules found in the human body including peptides, lipids, amino acids, nucleic acids, carbohydrates, organic acids, biogenic amines, vitamins, minerals, food additives, drugs, cosmetics, contaminants, pollutants, and just about any other chemical that humans ingest, metabolize, catabolize or come into contact with (1). Critical to the development of metabolomics (i.e. the study of metabolomes) has been the development of metabolomic databases (2). Just as genomics and proteomics depend on reference sequences in GenBank (3) and UniProt (4) to annotate genes and proteins, metabolomics critically depends on having reference compound data and reference spectral data to annotate metabolites. While many ‘metabolism’ databases exist, such as KEGG (5), Reactome (6) and the Cyc databases (7), relatively few true ‘metabolomics’ databases exist. Some of the better-known metabolomics databases include Metlin (8), MetaboLights (9), the Metabolomics Workbench (10), Lipid Maps (11) and the HMDB (12).
First released in 2007, the HMDB was one of the first and is currently the world's largest and most comprehensive, organism-specific metabolomic database. Designed to facilitate metabolomics research, the HMDB contains comprehensive, quantitative information about human metabolites along with their chemical structures, biological roles, physiological concentrations, disease associations, chemical reactions, transport mechanisms, metabolic pathways, and reference MS/MS (tandem mass spectrometry), GC–MS (gas chromatography mass spectrometry), and NMR (nuclear magnetic resonance) spectra. As with any primary data resource, the HMDB has continued to grow and evolve with the changing needs of its rapidly expanding user base, the advancing technologies and standards in metabolomics, and the continuing modernizations in web design and data delivery.
HMDB 1.0, introduced in 2007, provided a modest amount of biological, physiological and physico-chemical data on just 2180 human metabolites characterized and annotated as part of the Human Metabolome Project (13). HMDB 2.0, released in 2009, added much more referential (esp. NMR) spectral data and additional literature-derived molecular biological data for 6408 human metabolites (14). HMDB 3.0, released in 2013, added substantially more lipid and food constituent data, greatly expanded the HMDB’s spectral reference library, introduced far more metabolic pathway data and significantly modernized the HMDB user interface. HMDB 3.0 contained a total of 40 153 metabolites (15). While each of these prior HMDB releases provided notable improvements and greatly enriched the HMDB’s data content, this year's release represents the most significant expansion of the HMDB in its history.
This expansion has been motivated by an emerging crisis in metabolomics – a severe bottleneck in metabolite identification. In particular, despite more than two decades of metabolomics research, the proportion of human metabolites that can be identified via untargeted mass spectrometry (MS)-based metabolomics techniques is typically <2% of identified MS peaks (16). Furthermore, <10% of known human metabolites have authentic, experimentally collected NMR, GC–MS, or MS/MS spectra (15). These data suggest that existing chemical compound data sets and existing experimental spectral data sets for metabolomics are far too inadequate for comprehensive metabolite identification or quantification. Experimental solutions to this problem have been predicted to cost billions of dollars and to take decades of research (17). A more viable solution to this dilemma is to use in silico approaches to predict physiological feasible metabolites and to accurately predict NMR, GC–MS, or MS/MS spectra of both known and predicted metabolite structures. Fortunately, a number of very accurate open access and commercial tools for in silico metabolite and spectral prediction have recently become available (18–20).
Using these in silico approaches along with more traditional manual curation, the HMDB team has been able to expand the number of metabolites (known, expected and predicted) in HMDB 4.0 from 40 153 to 114 100 (nearly a threefold increase). In addition, the number of experimentally measured and computationally predicted NMR, MS/MS, and GC–MS reference spectra in HMDB 4.0 has grown from 3523 to 351 754 (nearly a 100-fold increase), the number of illustrated metabolic pathways in HMDB 4.0 has grown from 442 to more than 25,770 (nearly a 60-fold increase) and the number of metabolite-disease associations has increased from 3105 to 5498 (a 77% increase). In addition to this very significant expansion of its existing data, HMDB 4.0 has added many new data sets or novel kinds of data. This new data includes 18 192 metabolic reactions, new data on the influence of drugs on metabolite levels (pharmacometabolomics), the effect of SNP variations on metabolite levels in various biofluids, and the creation of a more consistent, fully defined metabolite ontology consisting of 4 categories, 35 subcategories and 3150 descriptors. Additionally, the HMDB curation team has greatly improved the quality and consistency of all existing metabolite descriptions, filled in data gaps on thousands of metabolite data entries, and greatly improved the quality and consistency of many chemical structure (esp. lipid) renderings. Major improvements to the spectral viewing and spectral search tools, spectral data formats (compatible with SPLASH (21), mzML (22) and nmrML (www.nmrml.org)), chemical taxonomies and chemical ontologies (23), and text and structure searching/matching have also been made.
Details regarding the HMDB’s overall layout, navigation structure, data sources, curation protocols, data management system, and quality assurance criteria have been described previously (14,15). This manuscript focuses on the additions and enhancements made to create HMDB 4.0.
DATABASE ADDITIONS AND IMPROVEMENTS
This section describes the three main areas where HMDB 4.0 has been significantly enhanced or improved. These include: (i) the expansion and enhancement of HMDB’s existing data, (ii) the addition of new data content and new data fields to HMDB 4.0, and (iii) improvements to the HMDB 4.0 interface.
Enhancement and expansion of existing data
Since 2007 the HMDB has seen a rapid expansion in the depth and breadth of its data as well as a significant enhancement to the quality and reliability of its information. The changes over the past 10 years are summarized in Table 1. The most noticeable change has been with the number of metabolites in the HMDB. Compared to the previous release (HMDB 3.0), HMDB 4.0 has greatly increased the total number of metabolites from 40 153 in HMDB 3.0 to 114 100 for HMDB 4.0. This corresponds to a nearly fivefold increase. As described in 2013, the HMDB grouped metabolites into three major classes: (i) Detected and quantified, (ii) Detected but not quantified, and (iii) Expected. Compounds classified as ‘Detected’ metabolites are those with measured concentrations or experimental confirmation of their existence in human biofluids, cells, or tissues. ‘Expected’ metabolites are those compounds of known structure for which biochemical pathways are known and where human intake/exposure is frequent, but the compound has yet to be detected in the body or the precise isomer has yet to be formally identified. For this year's release, the number of ‘Detected and quantified’ compounds increased slightly from 16 714 to 18 557 and the number of ‘Detected but not quantified’ compounds grew from 2798 to 3271. However, the category of compounds experiencing the largest growth has been the ‘Expected’ compounds, which grew from 20 641 compounds in HMDB 3.0 to 82 274 compounds in HMDB 4.0. Interestingly, the vast majority (>90%) of newly added ‘Expected’ metabolites are lipids. Because lipids are modular molecules with well-known biosynthetic pathways, head groups, and acyl/alkyl chain constituents, it is relatively easy to generate biologically feasible lipid structures. However, current MS and NMR technologies do not readily permit the identification of which acyl/alkyl chains are attached to which head group positions or where the saturated/desaturated bonds exist within these alkyl/acyl chains (17). As a result, the experimental evidence for specific lipid compounds is often lacking while the experimental evidence of specific lipid classes is often overwhelming. The inclusion of ‘Expected’ metabolites in 2013 was intended to help clear the metabolite identification bottleneck in metabolomics, however it only had a minor positive effect in clearing this bottleneck (as evidenced by the small increase of detected compounds in HMDB 4.0). Therefore for HMDB 4.0, a fourth class of metabolites (‘Predicted’) has been added. This new data category, its method of generation, and its rationale for inclusion will be discussed in more detail in the ‘New Data Content’ section.
Table 1. Comparison between the coverage in HMDB 1.0, 2.0, 3.0 and HMDB 4.0.
| Category | HMDB 1.0 | HMDB 2.0 | HMDB 3.0 | HMDB 4.0 |
|---|---|---|---|---|
| Total number of metabolites | 2180 | 6408 | 40 153 | 114 100 |
| Number of detected & quantified metabolites | 883 | 4413 | 16 714 | 18 557 |
| Number of detected, not quantified metabolites | 1297 | 1995 | 2798 | 3271 |
| Number of expected metabolites | 0 | 0 | 20 641 | 82 274 |
| Number of predicted metabolites* | 0 | 0 | 0 | 9548 |
| Number of unique synonyms | 27 700 | 43 882 | 199 668 | 1 231 398 |
| Number of cmpds with expt. MS/MS spectra | 390 | 799 | 1249 | 2265 |
| Number of cmpds with expt. GC/MS spectra | 0 | 279 | 1220 | 2544 |
| Number of cmpds with expt. NMR spectra | 385 | 792 | 1054 | 1494 |
| Number of cmpds with pred. MS/MS spectra* | 0 | 0 | 0 | 98 601 |
| Number of cmpds with pred. GC/MS spectra* | 0 | 0 | 0 | 26 880 |
| Number of experimental NMR spectra | 765 | 1580 | 2032 | 3840 |
| Number of experimental MS/MS spectra | 1180 | 2397 | 5776 | 22 198 |
| Number of experimental GC/MS spectra | 0 | 279 | 1763 | 7418 |
| Number of predicted MS/MS spectra* | 0 | 0 | 0 | 279 972 |
| Number of predicted GC/MS spectra* | 0 | 0 | 0 | 38 277 |
| Number of metabolic pathway maps | 26 | 58 | 442 | 25 570 |
| Number of compounds with disease links | 862 | 1002 | 3105 | 5498 |
| Number of compounds with concentration data | 883 | 4413 | 5027 | 7552 |
| Number of predicted molecular properties | 2 | 2 | 10 | 24 |
| Number of metabolite-SNP interactions* | 0 | 0 | 0 | 6777 |
| Number of metabolite-drug interactions* | 0 | 0 | 0 | 2497 |
| No. of metabolites w. sex/diurnal/age variation* | 0 | 0 | 0 | 2901 |
| Number of metabolic reactions* | 0 | 0 | 0 | 18,192 |
| Number of defined ontology terms* | 0 | 0 | 0 | 3150 |
| Number of HMDB data fields | 91 | 102 | 114 | 130 |
* New for HMDB 4.0
The past five years have seen an enormous growth in the availability of public, referential MS/MS, GC–MS, and NMR spectra of pure compounds. Many of these spectra can be found in resources such as MassBank of North America (MoNA), the GNPS (24), the Sumner lab MS library (25), MassBank (26), BioMagResBank (27) and others. At the same time, the HMDB curation team has also been actively collecting referential MS/MS, GC/MS, and NMR spectra using its own chemical libraries, MS and NMR instruments. In an effort to bring together the highly scattered public spectral data resources and to raise them to the same level of quality as found in the HMDB’s internally collected spectral libraries, the HMDB team has conducted an extensive spectral consolidation and remediation of publicly available MS and NMR data. Only experimentally collected, high-resolution (QTOF, Orbitrap or other) MS/MS and GC–MS spectra with detailed collection conditions and collision energy information have been included in the HMDB 4.0 release. Likewise, only high-resolution (400 MHz and greater) 1H or 13C NMR spectra have been collected and archived for HMDB 4.0. All MS/MS and GC–MS spectra have had their peaks annotated and fragment ions identified via CFM-ID (19) while all NMR spectra have been manually annotated with the assistance of commercial NMR spectral analysis programs (MNova and ACDLabs). Through this large-scale spectral consolidation effort, the number of experimentally measured MS/MS spectra in HMDB 4.0 has grown from 5776 (for 1249 compounds) in HMDB 3.0 to 22 247 (for 2265 compounds), which is a fourfold increase. Likewise the number of experimentally measured GC–MS spectra has grown from 1763 (for 1220 compounds) in HMDB 3.0 to 7418 (for 2544 compounds) for HMDB 4.0, a 300% increase. Finally, the number of experimentally measured NMR spectra has grown from 2032 (for 1054 compounds) in HMDB 3.0 to 3840 (for 1494 compounds) for HMDB 4.0, an 89% increase.
With HMDB 4.0, the quantity and quality of richly illustrated pathway diagrams has grown enormously. Previously, most of the pathway data in the HMDB was drawn from version 1.0 of SMPDB (28). Pathways from this early version of SMPDB were presented in the form of static images rendered using hand-drawn, manually hyperlinked powerpoint slides. In 2014/2015 the HMDB curation team developed a brand new tool called PathWhiz (29) for web-based pathway rendering and viewing. Using PathWhiz and its unique capacity to automatically ‘propagate’ manually generated template pathways, the HMDB curation team was able to substantially expand and update the content in SMPDB 2.0 (30), which is now fully linked to HMDB. Thanks to these efforts and the ongoing work to expand SMPDB, the total number of illustrated metabolic pathways in HMDB 4.0 has grown from 442 (in HMDB 3.0) to 25 570. Many (>95%) of these newly added pathways describe the catabolism/anabolism of lipids. The HMDB pathways have been grouped into several large categories: (i) metabolism/catabolism, (ii) metabolite-disease, (iii) metabolite-physiology, (iv) drug action, (v) drug metabolism, and (vi) metabolic signaling pathways. For HMDB 4.0, the number of metabolism/catabolism pathways is 25 086, the number of metabolite-disease pathways is 213, the number of metabolite-physiology pathways is 6, the number of drug action pathways is 383, the number of drug metabolism pathways is 64 and the number of metabolite signaling pathways is 18.
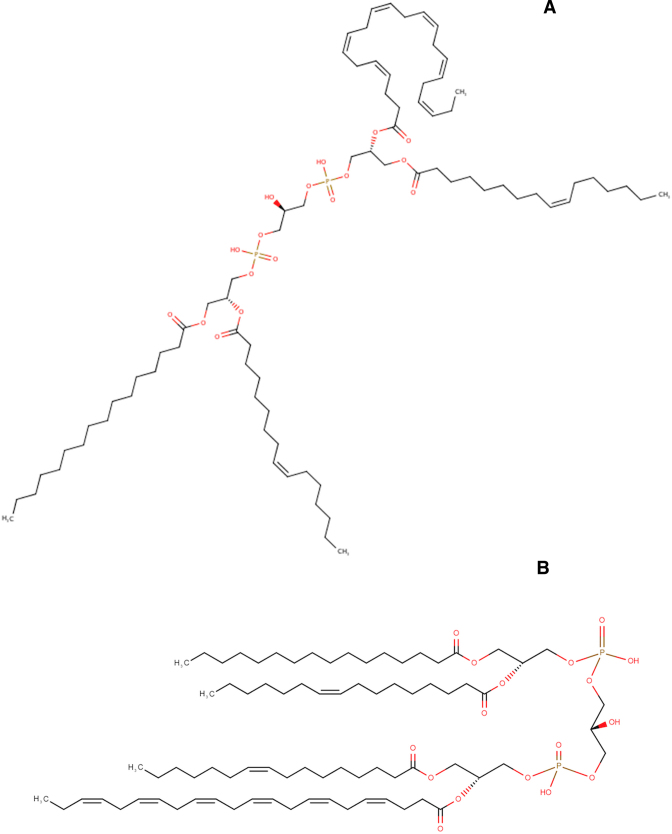
Another major effort taken on by the HMDB curation team has been structure remediation. Structure remediation involves the standardization, correction or re-rendering of stereoisomers, the removal/correction of salt forms, the regularization of how charged group are displayed and the standardization of how certain molecular entities (amines, carboxylates, tertiary amines, aromatic groups, protonation states, etc.) should be shown. Some of these corrections were done manually (via comparisons to other common rendering methods and other databases such as ChEBI (31) and PubChem (32)) while others were done automatically using specially developed in-house programs. For instance, many of the lipid structures in earlier versions of HMDB (esp. HMDB 3.0) were drawn using commercial software packages that generated lipid structures with a ‘squashed bug’ appearance (see Figure 1). In other words, the acyl chains of the lipid structures were randomly scattered using a naïve chain-avoidance algorithm. Multiple commercial chemical rendering programs were investigated to resolve this aesthetic issue, but without success. In the end, an in-house program was developed called ‘MetBuilder’ which generates lipid structures with all acyl/alkyl chains uniformly oriented in a parallel direction. The difference between the two types of lipid structure rendering can be seen in Figure 1 for the Cardiolipin(16:0/16:1/16:1/22:6). Obviously, the generation of lipid structures in this manner creates images that are more visually pleasing and more easily viewed or pasted into figures and slides. However, it is important to note that these pleasant-looking lipid structures are highly strained and the resulting MOL, SDF, and PDB files must be properly minimized if these files are to be used in any structural studies. Altogether more than 22,000 structures have been remediated or re-rendered in HMDB 4.0.
Figure 1.
An illustration of how the lipid structures looked like in HMDB 3.0 (A) and how they now appear in HMDB 4.0 (B) for the compound Cardiolipin(16:0/16:1/16:1/22:6).
Many biochemists and metabolomic researchers prefer to group metabolites according to their chemical classes (amino acids, lipids, carbohydrates). In early versions of the HMDB our curators manually classified metabolites into these standard classes. However, this approach was prone to errors and inconsistencies, especially when compounds were classified according to more traditional biological roles (i.e. vitamins) or when compounds had multiple chemical functionalities (sugar-lipids). In HMDB 3.0 a more automated, hierarchical chemical classification scheme (called ClassyFire) was introduced that was based on chemical structure alone. While this greatly improved consistency and reduced misclassification errors, the method was still in its infancy in 2012 and it lacked a formal ontology. Furthermore, it was not able to handle all structure inputs nor was it fully mapped or easily mapped to other chemical classification schemes used by ChEBI, LipidMaps, or PubChem (MeSH). With the finalization of the ClassyFire chemical classification system (23) and its chemical ontology (ChemOnt) in 2016, all 114,100 compounds within HMDB 4.0 have now been reclassified and updated to comply with the latest ClassyFire/ChemOnt release. Furthermore, the taxonomic mappings to other chemical classification schemes in other databases is now largely complete. More importantly, the ClassyFire classification scheme (which is fully open access) is now used by most other chemical/metabolomic databases (23). This means that the metabolomics community (and the chemical/biochemical community) now has a consistent, fully exchangeable standard for chemical classification.
In addition to substantially increasing the number of metabolites, spectra, and pathways in HMDB 4.0 as well as improving the quality of the structures and the chemical taxonomy in the database, our curation team has also added to the HMDB in many more ways. For instance, there are now many more compound names and synonyms (a 370% increase), substantially more compounds with biofluid concentration or tissue location data (a 50% increase), more data on normal and diseased metabolite concentrations (a 72% increase), and more data on biofluid and excreta metabolomes, including new data on urine (33), saliva (34), sweat, and feces. In particular, the number of metabolites (expected and detected) in human serum, urine, cerebrospinal fluid, saliva, sweat, and feces now stands at 25 424, 4225, 440, 1234, 90 and 1170, respectively.
Data quality and data currency continue to be two of the top priorities for the HMDB curation team. Because new metabolites from different tissues and biofluids are constantly being identified, re-characterized, re-measured or rescinded, the HMDB curation team continuously combs the literature to update existing MetaboCards (the data files associated with each metabolite in the HMDB) or to add new MetaboCards. As a result, thousands of metabolite descriptions, disease associations, biofluid/tissue concentrations, and metabolism data fields have been added, removed, re-written and/or expanded over the past five years.
New data fields and data content
As noted earlier, HMDB 4.0 now includes an entirely new class of compounds called ‘Predicted’ metabolites. This has been done in response to the growing metabolite identification crisis in metabolomics. Typically, fewer than 2% of confidently identified MS peaks in most metabolomic studies are being identified, even via simple mass matching (16,17). That number has largely remained unchanged over the past five years. It is clear that today's chemical databases (including the HMDB) are not providing the necessary metabolite coverage to allow researchers to identify these unknown or unidentifiable compounds. While a good deal of thought has gone into speculating what this metabolic ‘dark matter’ (16) might be, a growing consensus is that these unknowns are actually metabolites-of-metabolites (17). That is, these mystery compounds are enzymatically and non-enzymatically transformed products derived from well-known endogenous (or exogenous) compounds. In other words, metabolites-of-metabolites are commonly occurring chemicals in the body that have undergone phase I or phase II reactions in the liver, microbial transformations in the gut, or promiscuous enzyme reactions anywhere else in the body.
In silico metabolite prediction is a well-developed field of cheminformatics that has been central to advancing drug research and development for more than three decades (17). More recently, in silico metabolite modeling has been extended to the prediction of environmental degradation products through the EWAG-BBD System (18). Building from these well-developed ideas and precedents, the HMDB curation team has developed an in silico, open access metabolite prediction system called BioTransformer (Djoumbou Feunang, Y. (2017) Cheminformatics tools for enabling metabolomics. PhD Thesis, University of Alberta, Edmonton, AB, Canada). Given an input structure, BioTransformer predicts the site of metabolism, the reacting enzyme, and the resulting product structure(s) for single-step, multi-step more or multi-compartmental phase I, phase II, microbial and promiscuous enzymatic reactions. Extensive testing indicates that BioTransformer is able to predict biotransformation products with a precision of 46%, and a recall of 66%. It is also very good at identifying which compounds are NOT biotransformed (unlike most other in silico predictors). While still imperfect, BioTransformer's performance is 20–30% better than any other system (commercial or non-commercial). BioTransformer has been applied to a selected set of ‘Detected’ compounds in the HMDB. As a result, there are now 9548 ‘Predicted’ metabolites in HMDB 4.0, which represents about 8% of the current collection of metabolites in the HMDB. Every ‘Predicted’ metabolite is as fully (or almost as fully) annotated as a ‘Detected’ or ‘Expected’ metabolite. In particular, all ‘Predicted’ metabolites have a name (and synonyms), all have a textual description indicating the parent compound, the biotransformation or biotransforming enzyme, and a description of the chemical structure generated via ChemOnt (23). Additional nomenclature, structural, physico-chemical data, spectral data and biological (enzyme and reaction) data is also provided. All of the predicted compounds in HMDB 4.0 are labeled as ‘Predicted’ and users may easily exclude or include these compounds through HMDB’s data filters as they wish. HMDB’s ‘Predicted’ compounds are intended to serve as guides to help with the identification or characterization of unknown compounds.
The substantial expansion of annotated metabolites in HMDB 4.0 has necessitated an important change to the HMDB’s original 5-digit accession numbers. Since its inception, the HMDB has used unique accession numbers consisting of a 4-letter prefix (HMDB) and a 5-number suffix. With the release of HMDB 4.0, the 5-digit accession numbers have had to be adjusted to a 7-digit suffix to maintain consistency. This has been accomplished by appending two zeroes after the 4-letter prefix and before each 5-number suffix (e.g. HMDB00001 becomes HMDB0000001). In order to minimize disruption in dependent applications, the original 5-digit accession numbers have become secondary accession numbers to their 7-digit counterparts. This ensures that access to metabolite entries via their URLs is preserved.
Generating thousands of physiologically feasible structures only addresses half of the problem of metabolite identification. In particular, chemical structures themselves are not observable—only their spectra or retention times are. Over the past three years the HMDB curation team has been actively developing software to accurately predict the MS/MS, and GC–MS spectra of chemicals based on their known structure. In particular, CFM-ID (19) and CFM-ID 2.0 (35) are two tools that we developed that are able to accurately predict ESI-MS/MS spectra (under multiple collision energies) and GC–MS spectra. For this year's release of HMDB 4.0 all ‘Predicted’ compounds as well as all ‘Detected’ and ‘Expected’ compounds without existing experimental spectra have had their ESI-MS/MS spectra predicted (at collision energies of 10, 20 and 40 eV) with CFM-ID, their GC–MS spectra predicted (of appropriate TMS-derivatized compounds) with CFM-ID 2.0. In total, HMDB 4.0 contains 279 972 predicted MS/MS spectra for 98,601 compounds and 38,277 predicted GC–MS spectra for 26 880 compounds. All of the predicted MS spectra are labeled as ‘Predicted’ and they are intended to serve as guides to help with the identification or characterization of known or unknown compounds.
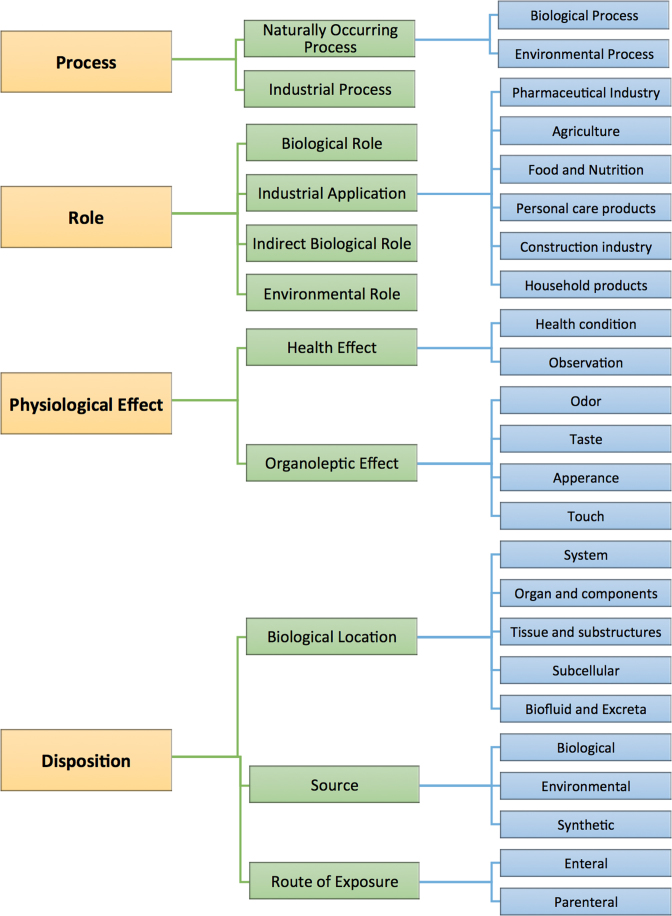
Another completely new set of data added to HMDB was a functional ontology. While ClassyFire (23) offers a very useful approach for structure-based classification and description, it does not solve all of the ontological problems with metabolomics or metabolites. In particular, the metabolomics community still needs a functional ontology similar to the Gene Ontology (GO) used in genomics and proteomics (36). In GO, genes and proteins are classified according to (i) cellular component, (ii) molecular function, and (iii) biological process. A modestly similar functional ontology was introduced in HMDB 3.0 covering (i) status (detected, expected), (ii) origin (exogenous/endogenous/microbial), (iii) biofunction, (iv) application, and (v) cellular location. However this early functional ontology did not use standard definitions or a standardized dictionary, and it only sparsely covered the metabolite ‘space’ in HMDB. In HMDB 4.0 a more sophisticated, hierarchical ontology has been introduced covering a more expansive set of terms. It also uses a fully standardized set of ontological definitions. In particular, the new HMDB ontology has four categories (process, role, physiological effect and disposition [biological location, source or route of exposure]), 35 subcategories (see Figure 2) and 3150 descriptors. All of the categories and descriptors are fully defined using standard English sentences and many terms have synonym sets to facilitate text mining. A more complete description of this chemical functional ontology (CFO) will be forthcoming in a future publication. All 114 100 compounds in the HMDB 4.0 have now been classified or partly classified using this new CFO. Many of the assignments were using a combination of manual annotation and a data mining software tools called PolySearch2 (37).
Figure 2.
A schematic diagram of the ontology structure in HMDB 4.0 for the first three ontological levels.
Many other new and significant additions to HMDB 4.0 have also been made. One of the more important ones includes the addition of pharmacometabolomics data. Pharmacometabolomics involves the analysis of how metabolite levels are changed in tissues, cells, or biofluids as a consequence of drug dosing (38). The pharmacometabolomic data was first mined from the literature and then manually curated and formatted so that users may identify which drugs increase or decrease metabolite levels in which tissues, cells, or biofluids. All drug names are hyperlinked to DrugBank (39) and all of the pharmacometabolomic data is browsable and searchable through a specially designed interface. In total, 764 drugs are linked to 494 metabolites through 2497 associations, which were compiled through 1317 publications. In addition to this pharmacometabolomic data, HMDB 4.0 also features a much richer collection of metabo-genomic or metabolite-SNP (single nucleotide polymorphism) data. With more and more metabolite-SNP studies being published each year, it is now clear that genetic mutation and polymorphisms do have a significant effect on baseline metabolite concentrations (40). While not as severe as the effects seen with inborn errors of metabolism, SNP-related metabolite effects can increase the risk for certain disorders. In total, HMDB 4.0 has 3056 metabolites linked to 2192 SNPs describing 6777 metabolite-SNP interactions. Metabolite levels are also known to differ with age, BMI, diurnal cycle, and gender. Given these effects, the HMDB now maintains a separate catalogue of 2901 metabolites that are known to co-vary with age, BMI, diurnal cycle, and gender. These data indicate which metabolites increase or decrease with these covariates and, where possible, by how much. Finally, a significant amount of new data is appearing in HMDB 4.0 concerning the ‘Exposome’ (41). The exposome is defined as the totality of chemical exposures that an individual may come into contact through daily living and along life course. These include food compounds, cosmetics, dyes, pesticides, pollutants, drugs, etc. All compounds that are considered to be part of the exposome are labeled as such and may be searched or categorized using HMDB’s Advanced Search engine.
New and enhanced interface features
Web technologies continue to advance and these advances can be exploited to make many web applications faster, more interesting, or more compelling. As always, the HMDB team continues to strive to improve the database's user interface, to enhance its search utilities, and to respond to user suggestions. For HMDB 4.0, we have introduced a number of interface improvements including (i) improved pathway viewing tools, (ii) improved spectral viewing/browsing tools, (iii) improved spectral searching, (iv) enhancing the advanced search, and (v) better resources for data exchange and interoperability.
HMDB 4.0 uses the PathWhiz system for pathway viewing. PathWhiz is a JavaScript enabled tool that allows users to interactively draw, view, zoom and click on colourful pathway images that are richly annotated with cellular organelle data, organ/tissue data, enzyme cofactor information, and protein quaternary structure information. All images are extensively hyperlinked (to UniProt, DrugBank, and HMDB) and all pathways are carefully described or summarized with an explanatory paragraph. To facilitate pathway rendering, two types of pathway views are now available including a (default) data rich pathway rendering, and a simplified KEGG-like pathway rendering which is more amenable for slides, figures, or manuscripts. All of the pathways rendered in HMDB can be saved and downloaded in static image formats (PNG and SVG) as well as in a variety of common data exchange formats such as SBML (systems biology mark-up language), BioPax, and PathWhiz's own markup language (PWML).
The large quantity of spectral data in HMDB precipitated a need to improve the HMDB’s spectral viewing tools. Previously static PDF images or relatively crude ‘stick’ renderings were the norm for most of HMDB’s spectral data. With HMDB 4.0 all MS and NMR spectra are now viewable with JSpectraViewer, a locally developed JavaScript spectral viewing tool that is able to read and render nmML and mzML formatted spectra. For NMR spectra, JSpectraViewer allows users to interactively hover over spectral peaks and to see 1H spectral assignments and the corresponding protons on a rendered image of the molecule. For MS spectra, JSpectraViewer allows users to interactively hover over spectral peaks and to see the structures of the fragment ions that are predicted (via CFM-ID) to correspond to the observed m/z value.
The improvements to the HMDB’s spectral viewing tools have also led to a number of improvements to HMDB’s spectral searching tools. In particular, HMDB’s MS spectral search and MS spectral results page has been redesigned to better reflect the needs and expectations of MS spectroscopists. In particular, the MS Search page has improved the way users can select adducts by reformatting and simplifying ‘Adduct Type’ field. In addition, an ‘m/z calculator’ has been introduced in MS Search results page to facilitate rapid comparisons and calculations and the page has been further modified to display more information including the chemical formula, the monoisotopic mass and the mass differences from the query mass (i.e. Delta(ppm)). We have also included KEGG and HMDB identifiers in the extractable CSV file for simplifying data analysis and data extraction. Furthermore, all MS spectra in HMDB are now mapped to a SPLASH (SPectraL hASH) key (21). This spectral hash identifier allows MS spectra within HMDB to be easily searched or identified via the web through a standard Google (or other search engine) query. With the continued additions of new data fields and growing requests for new kinds of queries from its users, the HMDB team has also been actively improving and correcting HMDB’s Advanced Search. The interface is more intuitive and powerful. The Advanced Search allows you to build structured queries containing predicates such as ‘matches/does not match’, ‘starts/ends with’, ‘greater/less than’, and ‘is/is not present’. It has been corrected to include negative value in search. The result interface now allows one to browse the results through pagination.
Because much of the data in the HMDB is downloadable and much of its data is being used to create or supplement other metabolomics resources, the need for improved data exchange standards and improved interoperability within the HMDB has become much greater. In response to these needs, all of HMDB’s chemical structures are now accessible in canonical SMILES, SDF, MOL, PDB, InChI, and InChIKey formats. Furthermore, all experimentally observed and theoretically predicted MS/MS, GC–MSand NMR spectra are stored and downloadable in mzML (22) and nmrML (www.nmrml.org) formats and all MS/MS and GC–MS spectra are assigned SPLASH keys (21) for rapid spectral querying and matching. Likewise, all sequence (DNA and protein) data is stored in FASTA format and all remaining textual data is stored in XML and JSON format. All of these HMDB ‘component’ files can be freely downloaded (for academics) from the HMDB 4.0 website.
CONCLUSION
Over the past 10 years the HMDB has grown from a small, somewhat specialized chemical database to the largest, most comprehensive, organism-specific metabolomic ‘knowledgebase’ in the world. Throughout this evolution we have strived to maintain the highest quality and currency of data, to continuously improve the quality of its user interface, and to attentively listen to all members of the metabolomics community. With this latest release of the HMDB we have undertaken the most significant update in its history. Many existing data sets have grown in size by factors of 3–30-fold. In addition to this significant enhancement to the HMDB’s existing data collection, we have also added substantial quantities of new and very important data for the entire metabolomics community. In particular, HMDB 4.0 now includes predicted metabolites, predicted NMR, GC–MS, and MS/MS spectral data as well as predicted retention time data. These data sets are intended to address the metabolite identification crisis, which is severely limiting compound coverage for much of today's metabolomic studies. Other kinds of ‘metabo-omic’ data such as pharmacometabolomic data and geno-metabolomic data have also been added to make the HMDB much more compatible with the increasing number of multi-omic or systems biology studies that are being undertaken. Other improvements to the HMDB’s interface, spectral and pathway viewing tools, searching utilities and data exchange standards should also make the HMDB much easier to work with and more enjoyable to explore. All of these additions and enhancements are intended to facilitate research in human nutrition, biochemistry, clinical chemistry, clinical genetics, medicine and metabolomics science.
ACKNOWLEDGEMENTS
The authors would like to thank the many users of the HMDB for their valuable feedback and suggestions.
FUNDING
Genome Alberta (a division of Genome Canada); The Canadian Institutes of Health Research (CIHR); Western Economic Diversification (WED); Alberta Innovates Health Solutions (AIHS). Funding for open access charge: Genome Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wishart D.S. Metabolomics: applications to food science and nutrition research. Trends Food Sci. Technol. 2008; 19:482–493. [Google Scholar]
- 2. Wishart D.S. Current progress in computational metabolomics. Brief. Bioinf. 2007; 8:279–293. [DOI] [PubMed] [Google Scholar]
- 3. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2017; 45:D37–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S. et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016; 44:D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016; 44:D471–D480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith C.A., O’Maille G., Want E.J., Qin C., Trauger S.A., Brandon T.R., Custodio D.E., Abagyan R., Siuzdak G.. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005; 27:747–751. [DOI] [PubMed] [Google Scholar]
- 9. Kale N.S., Haug K., Conesa P., Jayseelan K., Moreno P., Rocca-Serra P., Nainala V.C., Spicer R.A., Williams M., Li X. et al. MetaboLights: an open-access database repository for metabolomics data. Curr. Protoc. Bioinformatics. 2016; 53:14.13.1–14.13.18. [DOI] [PubMed] [Google Scholar]
- 10. Sud M., Fahy E., Cotter D., Azam K., Vadivelu I., Burant C., Edison A., Fiehn O., Higashi R., Nair K.S. et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016; 44:D463–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sud M., Fahy E., Cotter D., Brown A., Dennis E.A., Glass C.K., Merrill A.H. Jr, Murphy R.C., Raetz C.R., Russell D.W., Subramaniam S.. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007; 35:D527–D532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wishart D.S., Tzur D., Knox C., Eisner R., Guo A.C., Young N., Cheng D., Jewell K., Arndt D., Sawhney S. et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007; 35:D521–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wishart D.S. Proteomics and the human metabolome project. Expert Rev. Proteomics. 2007; 4:333–335. [DOI] [PubMed] [Google Scholar]
- 14. Wishart D.S., Knox C., Guo A.C., Eisner R., Young N., Gautam B., Hau D.D., Psychogios N., Dong E., Bouatra S. et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009; 37:D603–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wishart D.S., Jewison T., Guo A.C., Wilson M., Knox C., Liu Y., Djoumbou Y., Mandal R., Aziat F., Dong E. et al. HMDB 3.0—the Human Metabolome Database in 2013. Nucleic Acids Res. 41:D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. da Silva R.R., Dorrestein P.C., Quinn R.A.. Illuminating the dark matter in metabolomics. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:12549–12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dias D.A., Jones O.A., Beale D.J., Boughton B.A., Benheim D., Kouremenos K.A., Wolfender J.L., Wishart D.S.. Current and future perspectives on the structural identification of small molecules in biological systems. Metabolites. 2016; 6:E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao J., Ellis L.B., Wackett L.P.. The University of Minnesota Biocatalysis/Biodegradation Database: improving public access. Nucleic Acids Res. 2010; 38:D488–D491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Allen F., Pon A., Wilson M., Greiner R., Wishart D.. CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res. 2014; 42:W94–W99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeffryes J.G., Colastani R.L., Elbadawi-Sidhu M., Kind T., Niehaus T.D., Broadbelt L.J., Hanson A.D., Fiehn O., Tyo K.E., Henry C.S.. MINEs: open access databases of computationally predicted enzyme promiscuity products for untargeted metabolomics. J. Cheminform. 2015; 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wohlgemuth G., Mehta S.S., Mejia R.F., Neumann S., Pedrosa D., Pluskal T., Schymanski E.L., Willighagen E.L., Wilson M., Wishart D.S. et al. SPLASH, a hashed identifier for mass spectra. Nat. Biotechnol. 2016; 34:1099–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deutsch E. mzML: a single, unifying data format for mass spectrometer output. Proteomics. 2008; 8:2776–2777. [DOI] [PubMed] [Google Scholar]
- 23. Djoumbou Feunang Y., Eisner R., Knox C., Chepelev L., Hastings J., Owen G., Fahy E., Steinbeck C., Subramanian S., Bolton E. et al. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016; 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C.A., Luzzatto-Knaan T. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016; 34:828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lei Z., Jing L., Qiu F., Zhang H., Huhman D., Zhou Z., Sumner L.W.. Construction of an ultrahigh pressure liquid chromatography-tandem mass spectral library of plant natural products and comparative spectral analyses. Anal. Chem. 2015; 87:7373–7381. [DOI] [PubMed] [Google Scholar]
- 26. Horai H., Arita M., Kanaya S., Nihei Y., Ikeda T., Suwa K., Ojima Y., Tanaka K., Tanaka S., Aoshima K. et al. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010; 45:703–714. [DOI] [PubMed] [Google Scholar]
- 27. Ulrich E.L., Akutsu H., Doreleijers J.F., Harano Y., Ioannidis Y.E., Lin J., Livny M., Mading S., Maziuk D., Miller Z. et al. BioMagResBank. Nucleic Acids Res. 2008; 36:D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frolkis A., Knox C., Lim E., Jewison T., Law V., Hau D.D., Liu P., Gautam B., Ly S., Guo A.C. et al. SMPDB: the Small Molecule Pathway Database. Nucleic Acids Res. 2010; 38:D480–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pon A., Jewison T., Su Y., Liang Y., Knox C., Maciejewski A., Wilson M., Wishart D.S.. Pathways with PathWhiz. Nucleic Acids Res. 2015; 43:W552–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jewison T., Su Y., Disfany F.M., Liang Y., Knox C., Maciejewski A., Poelzer J., Huynh J., Zhou Y., Arndt D. et al. SMPDB 2.0: big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014; 42:D478–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C.. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016; 44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bouatra S., Aziat F., Mandal R., Guo A.C., Wilson M.R., Knox C., Bjorndahl T.C., Krishnamurthy R., Saleem F., Liu P. et al. The human urine metabolome. PLoS One. 2013; 8:e73076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dame Z.T., Aziat F., Mandal R., Krishnamurthy R., Bouatra S., Borzouie S., Guo A.C., Sajed T., Deng L., Lin H. et al. The Human Saliva Metabolome. Metabolomics. 2015; 11:1864–1883. [Google Scholar]
- 35. Allen F., Pon A., Greiner R., Wishart D.. Computational Prediction of Electron Ionization Mass Spectra to Assist in GC/MS Compound Identification. Anal. Chem. 2016; 88:7689–7697. [DOI] [PubMed] [Google Scholar]
- 36. The Gene Ontology Consortium Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017; 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y., Liang Y., Wishart D.S.. PolySearch2: a significantly improved text-mining system for discovering associations between human diseases, genes, drugs, metabolites, toxins and more. Nucleic Acids Res. 2015; 43:W535–W542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaddurah-Daouk R., Weinshilboum R. the Pharmacometabolomics Research Network . Metabolomic signatures for drug response phenotypes: pharmacometabolomics enables precision medicine. Clin. Pharmacol. Ther. 2015; 98:71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Law V., Knox C., Djoumbou Y., Jewison T., Guo A. C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014; 42:D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kastenmüller G., Raffler J., Gieger C., Suhre K.. Genetics of human metabolism: an update. Hum. Mol. Genet. 2015; 24:R93–R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Neveu V., Moussy A., Rouaix H., Wedekind R., Pon A., Knox C., Wishart D.S., Scalbert A.. Exposome-Explorer: a manually-curated database on biomarkers of exposure to dietary and environmental factors. Nucleic Acids Res. 2017; 45:D979–D984. [DOI] [PMC free article] [PubMed] [Google Scholar]