Abstract
CRISPR-Cas is a tool that is widely used for gene editing. However, unexpected off-target effects may occur as a result of long-term nuclease activity. Anti-CRISPR proteins, which are powerful molecules that inhibit the CRISPR–Cas system, may have the potential to promote better utilization of the CRISPR-Cas system in gene editing, especially for gene therapy. Additionally, more in-depth research on these proteins would help researchers to better understand the co-evolution of bacteria and phages. Therefore, it is necessary to collect and integrate data on various types of anti-CRISPRs. Herein, data on these proteins were manually gathered through data screening of the literatures. Then, the first online resource, anti-CRISPRdb, was constructed for effectively organizing these proteins. It contains the available protein sequences, DNA sequences, coding regions, source organisms, taxonomy, virulence, protein interactors and their corresponding three-dimensional structures. Users can access our database at http://cefg.uestc.edu.cn/anti-CRISPRdb/ without registration. We believe that the anti-CRISPRdb can be used as a resource to facilitate research on anti-CRISPR proteins and in related fields.
INTRODUCTION
Several types of defense mechanism exist in Archaea and bacteria to prevent invaders, such as phages and plasmids, from infecting the bodies of these organisms (1). Among the defense weapons, CRISPR-Cas system (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) constitutes a major and powerful adaptive immune system of bacteria that helps bacteria to escape the threat of viruses and other mobile genetic elements (MGEs) (2). Data stored in the CRISPRdb database demonstrates that CRISPR-Cas systems are widely present in bacteria and Archaea (3). Makarova et al. have provided a classification of CRISPR–Cas systems based on signature protein families and features of the architecture of Cas loci, and their classification contained two major classes that are divided into five types and 16 subtypes (4). Recently, three new subtypes of type V, and three subtypes of type VI were predicted by running a pipeline against all available bacterial, archaeal genomes and metagenomics contigs, respectively, and the activities of corresponding effectors have been validated by experiments (5–8). In 2013, Bondy-Denomy et al. first found anti-CRISPR proteins with anti-I-F activity in Pseudomonas aeruginosa phage (9). Their studies opened the door to investigate anti-CRISPR proteins. Anti-CRISPRs offer a powerful defense system that helps phages to escape injury from the CRISPR-Cas system. In order to escape the wide scope of the CRISPR–Cas system, phages may evolve various types of anti-CRISPRs. Following the discovery of the first type anti-CRISPRs, more anti-CRISPR proteins with anti-I-F, anti-I-E, anti-II-A, anti-II-C, and anti-VI-B activities were identified (5,10–13). In addition, the structures of particular anti-CRISPR proteins in complex with other proteins were resolved by the scientific community (14–17). Especially striking are three recent studies that reported the structure of SpyCas9–sgRNA–AcrIIA4 ternary complexes and showed that AcrIIA4 can inhibit SpyCas9 activity via obscuring the protospacer adjacent motif (PAM) on the one hand; on the other hand, AcrIIA4 can also shield the RuvC active site in SpyCas9 (18–20).
CRISPR-Cas technology is a tool that has been widely used for gene editing. However, off-target effects may appear as a result of long-term nuclease activity (21). Some methods such as GUIDE-Seq have been developed to detect the genome-wide profiling of off-target cleavage by CRISPR–Cas nucleases (22). The off-target phenomenon is unexpected when scientists perform gene editing. Anti-CRISPR proteins can be considered as powerful molecules that switch off the activity of CRISPR-Cas (12). The experiment performed by Shing et al. certified that AcrIIA4 has the potential to reduce off-target events in both research and therapeutic applications (19). Hence, the discovery of anti-CRISPR proteins offers promise for possibly solving off-target effects. In addition, the in-depth study of these proteins can help us to better understand the co-evolution between bacteria and phages. Recently, a review has systematically introduced those anti-CRISPR proteins from the aspect of their discovery, mechanisms, and evolutionary impact, which can help the scientific community further understand them (23).
To our knowledge, no online resource for organizing these data is available to date. Therefore, it is necessary to collect and integrate data on various types of anti-CRISPR proteins. To this end, we gathered data on all of these proteins through data screening using the literatures published since the first anti-CRISPR protein was identified and then constructed an online resource named anti-CRISPRdb. The first version of anti-CRISPRdb currently contains 432 anti-CRISPR proteins tested by experimental and bioinformatics methods. These proteins could be divided into the following five categories: anti-CRISPR proteins with anti-I-F activity, those with anti-I-E activity, those with anti-II-A activity, those with anti-II-C activity, and those with anti-VI-B activity. Furthermore, we also provided a non-redundant dataset that contains 106 sequences. Our database can be freely accessed at http://cefg.uestc.edu.cn/anti-CRISPRdb/ without the need for registration.
DATA SOURCE AND DATABASE CONSTRUCTION
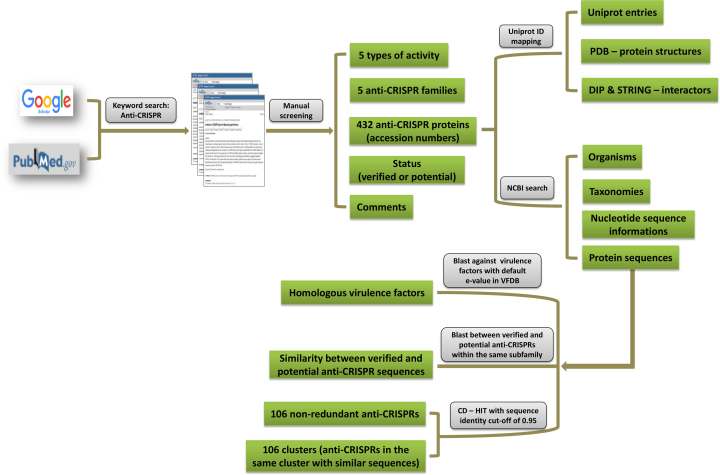
Figure 1 illustrates the construction process of anti-CRISPRdb. First, we obtained related references regarding anti-CRISPR proteins using keywords for the retrieval of papers in PubMed and Google Scholar. Then, we screened the information associated with anti-CRISPR proteins from those references, such as protein accession IDs, anti-CRISPR types, and pivotal comments and families. According to protein accession numbers, Python script (version 2.7) was used to download protein information, including protein sequences, corresponding nucleotide sequences, coding regions, source organisms, and taxonomy from NCBI (National Center for Biotechnology Information). Information about the similarity of potential and verified anti-CRISPRs is valuable because it can help us understand how similar a putative anti-CRISPR is to the most closely verified anti-CRISPR. Therefore, we aligned each pair of sequences within the same sub-family using BLAST 2.2.30+ with default parameters and provided this information in the database. Additionally, these anti-CRISPR proteins should interact with other molecules and form specific spatial structures to carry out their functions. Therefore, we also collected structure and interaction information from the PDB (Protein Data Bank), STRING (Search Tool for Recurring Instances of Neighboring Genes) and DIP (Database of Interacting Proteins) databases via ID mapping for each anti-CRISPR (24–26). VFDB (Virulence Factors Database) is an comprehensive database that curates information on the virulence factors (VFs) of bacterial pathogens (27). In addition to high anti-CRISPR activity, an ideal anti-CRISPR protein should have a lower cytotoxicity for cells that we want to reduce off-target sites. For this purpose, we conducted a BLASTp search against the VFDB_setB_pro.fas dataset downloaded from VFDB, and provided homologous information about anti-CRISPR proteins and virulence factors, which can help users to explore the cytotoxicity of anti-CRISPRs. Many of the anti-CRISPR protein orthologs in closely related bacterial strains are 100% identical. Therefore, we used the CD-HIT web server (http://weizhongli-lab.org/cdhit_suite/cgi-bin/index.cgi?cmd=cd-hit) to screen non-redundant sequences with a cut-off value of 95% (28). The anti-CRISPR proteins were grouped into 106 clusters by CD-HIT, and each group was represented by one representative sequence. Users can access this non-redundant dataset in our database via a specially designed web interface. Moreover, users can browse members in each cluster and the similarity between each member and the representative sequence.
Figure 1.
The flow chart illustrates the construction process of anti-CRISPRdb, and the information that users can obtain from anti-CRISPRdb.
Our database anti-CRISPRdb adopts the framework of LAMP, which is a popular web architecture that contains a Linux system, an Apache Network Server, the MySQL database, and PHP/Python programming language, and runs with the Ubuntu 10.04. We tested our database using different browsers on different systems (IE, Firefox and Chrome browsers on a Windows operating system; Firefox and Chromium browsers on a Linux operating system; and Safari and Chrome browsers on a Mac operating system) to ensure normal display. The testing results exhibited good compatibility.
HOW TO USE anti-CRISPRdb
The main, internal and external pages of anti-CRISPRdb
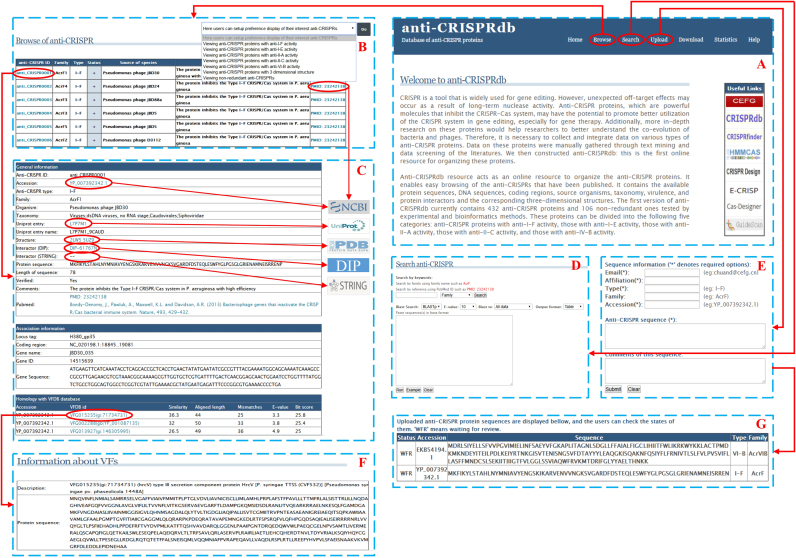
Figure 2 is a screenshot of the anti-CRISPRdb web interface that illustrates the relationship between the internal and external pages. Figure 2A represents the home page; and Figure 2B represents the browser page. Upon clicking any anti-CRISPR ID, detailed information for each anti-CRISPR protein is displayed in Figure 2C. Figure 2C describes detailed information about each anti-CRISPR protein. Here, users can obtain information regarding protein annotations (such as the source organism, taxonomy, and activity), protein structures, and their corresponding interactors. On this page, we provided links to the NCBI, Uniprot, PDB, STRING, and DIP databases, which can help users to further explore anti-CRISPR proteins. Figure 2D is a search page. Figure 2E is a page designed for uploading data on anti-CRISPR proteins, and Figure 2G lists the proteins that are uploaded by users. Figure 2F shows information about virulence factors.
Figure 2.
A screenshot of the anti-CRISPRdb web interface, which can illustrate the relationship among the main, internal and external pages. Please note data showing in Figure 2G is an example.
The main functions of anti-CRISPRdb
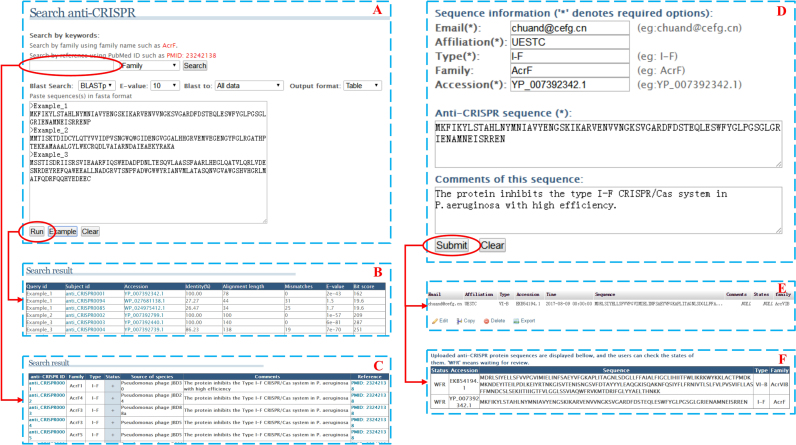
In our database, we provide useful functions for searching, browsing, downloading, and uploading data on anti-CRISPR proteins, which can facilitate researchers’ use of the database. On the browser page, we provided important information such as family, function, source species, verification status, comments, and related references. In addition, anti-CRISPR proteins with known structures and different types can also be accessed in this browser interface. We also designed an interface so that users can easily access the non-redundant dataset. On the search page, users can search for anti-CRISPRs of interest using the family name, accession number, source organism, anti-CRISPR ID, and reference ID. This search function can also function as an API (Application Programming Interface). We also integrated the BLAST program into the anti-CRISPR database, which can help researchers to discover potential anti-CRISPR proteins based on input proteins of interest via alignment with proteins in the database (29). Figure 3 shows an example demonstrating how to use the search function. Multi-sequences in FASTA format can be pasted into the box. Users perform the BLAST search program after setting their preferred parameters or maintaining the default parameters (Figure 3A). To present a clear visual display, the alignment and search results are displayed using a table format by default (Figure 3B and C). By clicking any of the IDs in parts Figure 3B and C, users can obtain more detailed information. Because the pairwise alignment output can be helpful for validating the BLAST results in some cases, search results can also be displayed in a pairwise alignment format. Blast results that have high bit-score will be preferentially displayed. The configurable BLAST service can help the users to further screen the alignments.
Figure 3.
This is an example demonstrating how to use the search and upload interfaces. Please note data showing in Figure 3F is an example.
In addition to the free data accessibility for an open-access database, data on validated/potential anti-CRISPR proteins identified by other scientific communities can be uploaded. This setting can facilitate the study of anti-CRISPR proteins. For this feature, we designed an interface that can allow users to share the latest data on newly identified anti-CRISPR proteins. Figure 3 also illustrates how to share data on anti-CRISPR proteins with the related scientific community. For the upload interface (Figure 3D), there are five fields that uploaders must fill in: email, the affiliation of the users, anti-CRISPR type, protein accession number, and protein sequence. Sequence information will be uploaded into the backstage (Figure 3E) for the administrator's review after the submit button has been pressed, and sequence information can be displayed in Figure 3F without showing the uploader's information.
DISCUSSION
CRISPR-Cas technology has been widely used in various applications, such as therapeutic application for HIV (30), biomedical discoveries (31), and essential gene screening (32,33). However, undesirable off-target effects can limit editing effectiveness and safety. It is critical to pre-detect off-target sites in a given genome in order to design ‘off-switch’ tools. Some methods to assess potential off-target sites have been developed, such as CasOT, which is an exhaustive tool that can be used to precisely identify potential off-target sites in the genome and other sequences (34). To guarantee increased safety when using CRISPR–Cas9 technology in gene therapy, we should be able to turn off CRISPR activity in places where we do not want to perform editing at a particular time. In addition, anti-CRISPR proteins can inhibit the function of the CRISPR–Cas system, and can thereby help researchers to better utilize CRISPR-Cas technology. Given the crucial significance of this system, we constructed an online resource anti-CRISPRdb to integrate data on these proteins. Users can not only browse, search, blast, screen, and download data on their anti-CRISPR proteins of interest, but can also share data on validated/potential anti-CRISPR proteins with other related scientific communities. Hence, our database is a user-friendly database that encourages data sharing.
It has been a short time since the first anti-CRISPR protein was identified. Since that time, only five types (anti-I-F, anti-I-E, anti-II-A, anti-II-C and anti-VI-B) of anti-CRISPRs have been discovered. However, the wide spread existence of the CRISPR-Cas system implies that other types of anti-CRISPR protein exist (owing to the co-evolution of bacteria and phages). With progress in the technologies for identifying anti-CRISPR proteins, new types will be gradually identified, and our database will be updated regularly.
ACKNOWLEDGEMENTS
We would like to thank Dr Alan R. Davidson who kindly provided us with four anti-CRISPR protein sequences that have anti-I-E activity, and we thank Dr Jian Yang, who kindly replied our questions about the VFDB database. Additionally, we thank the three anonymous reviewers’ constructive comments and suggestions.
FUNDING
National Natural Science Foundation of China [31470068, 81171411]; Sichuan Province Science and Technology Support Plan [2015SZ0191]; Fundamental Research Funds for the Central Universities of China [ZYGX2016J117, ZYGX2015Z006, ZYGX2015J144]. Funding for open access charge: National Natural Science Foundation of China [31470068, 81171411].
Conflict of interest statement. None declared.
REFERENCES
- 1. Dupuis M.E., Villion M., Magadan A.H., Moineau S.. CRISPR–Cas and restriction-modification systems are compatible and increase phage resistance. Nat. Commun. 2013; 4:2087. [DOI] [PubMed] [Google Scholar]
- 2. Horvath P., Barrangou R.. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010; 327:167–170. [DOI] [PubMed] [Google Scholar]
- 3. Grissa I., Vergnaud G., Pourcel C.. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007; 8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smargon A.A., Cox D.B., Pyzocha N.K., Zheng K., Slaymaker I.M., Gootenberg J.S., Abudayyeh O.A., Essletzbichler P., Shmakov S., Makarova K.S.. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell. 2017; 65:618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koonin E.V., Makarova K.S., Zhang F.. Diversity, classification and evolution of CRISPR–Cas systems. Curr. Opin. Microbiol. 2017; 37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K.. Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell. 2015; 60:385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L.. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016; 353:aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bondy-Denomy J., Pawluk A., Maxwell K.L., Davidson A.R.. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013; 493:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pawluk A., Bondy-Denomy J., Cheung V.H., Maxwell K.L., Davidson A.R.. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR–Cas system of Pseudomonas aeruginosa. mBio. 2014; 5:e00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pawluk A., Staals R.H., Taylor C., Watson B.N., Saha S., Fineran P.C., Maxwell K.L., Davidson A.R.. Inactivation of CRISPR–Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 2016; 1:16085. [DOI] [PubMed] [Google Scholar]
- 12. Pawluk A., Amrani N., Zhang Y., Garcia B., Hidalgo-Reyes Y., Lee J., Edraki A., Shah M., Sontheimer E.J., Maxwell K.L. et al. Naturally occurring off-switches for CRISPR–Cas9. Cell. 2016; 167:1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rauch B.J., Silvis M.R., Hultquist J.F., Waters C.S., McGregor M.J., Krogan N.J., Bondy-Denomy J.. Inhibition of CRISPR–Cas9 with bacteriophage proteins. Cell. 2017; 168:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maxwell K.L., Garcia B., Bondy-Denomy J., Bona D., Hidalgo-Reyes Y., Davidson A.R.. The solution structure of an anti-CRISPR protein. Nat. Commun. 2016; 7:13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chowdhury S., Carter J., Rollins M.F., Golden S.M., Jackson R.N., Hoffmann C., Bondy-Denomy J., Maxwell K.L., Davidson A.R., Fischer E.R.. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell. 2017; 169:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X., Yao D., Xu J.G., Li A.-R., Xu J., Fu P., Zhou Y., Zhu Y.. Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat. Struct. Mol. Biol. 2016; 23:868–870. [DOI] [PubMed] [Google Scholar]
- 17. Wang J., Ma J., Cheng Z., Meng X., You L., Wang M., Zhang X., Wang Y.. A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses. Cell Res. 2016; 26:1165–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dong D., Guo M., Wang S., Zhu Y., Wang S., Xiong Z., Yang J., Xu Z., Huang Z.. Structural basis of CRISPR–SpyCas9 inhibition by an anti-CRISPR protein. Nature. 2017; 546:436–439. [DOI] [PubMed] [Google Scholar]
- 19. Shing J., Jiang F., Liu J.J., Bray N.L., Rauch B.J., Baik S.H., Nogales E., Bondy-Denomy J., Corn J.E., Doudna J.A.. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 2017; 3:e1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang H., Patel D.J.. Inhibition mechanism of an anti-CRISPR suppressor AcrIIA4 targeting SpyCas9. Mol. Cell. 2017; 67:117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR–Cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P.. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR–Cas nucleases. Nat. Biotechnol. 2015; 33:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Borges A.L., Davidson A.R., Bondy-Denomy J.. The discovery, mechanisms, and evolutionary impact of anti-CRISPRs. Annu. Rev. Virol. 2017; 4, doi:10.1146/annurev-virology-101416-041616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sussman J.L., Lin D., Jiang J., Manning N.O., Prilusky J., Ritter O., Abola E.. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D. Biol. Crystallogr. 1998; 54:1078–1084. [DOI] [PubMed] [Google Scholar]
- 25. Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P.. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016; 45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D.. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004; 32:D449–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen L., Zheng D., Liu B., Yang J., Jin Q.. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016; 44:D694–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang Y., Niu B., Gao Y., Fu L., Li W.. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010; 26:680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shiryev S.A., Papadopoulos J.S., Schäffer A.A., Agarwala R.. Improved BLAST searches using longer words for protein seeding. Bioinformatics. 2007; 23:2949–2951. [DOI] [PubMed] [Google Scholar]
- 30. Saayman S., Ali S.A., Morris K.V., Weinberg M.S.. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opin. Biol. Ther. 2015; 15:819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Riordan S.M., Heruth D.P., Zhang L.Q., Ye S.Q.. Application of CRISPR/Cas9 for biomedical discoveries. Cell Biosci. 2015; 5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., Sabatini D.M.. Identification and characterization of essential genes in the human genome. Science. 2015; 350:1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S. et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015; 163:1515–1526. [DOI] [PubMed] [Google Scholar]
- 34. Xiao A., Cheng Z., Kong L., Zhu Z., Lin S., Gao G., Zhang B.. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics. 2014; 30:1180–1182. [DOI] [PubMed] [Google Scholar]