Abstract
Living cells are made up of individual parts, i.e. the genome, the proteins, the RNA and lipid molecules as well as the metabolites and ions. However, life depends on the functional interaction among these components which is often organized in networks. Here, we present the recent development of SubtiWiki, the integrated database for the model bacterium Bacillus subtilis (http://subtiwiki.uni-goettingen.de/). SubtiWiki is based on a relational database and provides access to published information about the genes and proteins of B. subtilis and about metabolic and regulatory pathways. We have included a network visualization tool that can be used to visualize regulatory as well as protein-protein interaction networks. The resulting interactive graphical presentations allow the user to detect novel associations and thus to develop novel hypotheses that can then be tested experimentally. To facilitate the mobile use of SubtiWiki, we provide enhanced versions of the SubtiWiki App that are available for iOS and Android devices. Importantly, the App allows to link private notes and pictures to the gene/protein pages that can be synchronized on multiple devices. SubtiWiki has become one of the most complete resources of knowledge on a living organism.
INTRODUCTION
Model organisms are essential to develop our knowledge in biology as they are in the focus of research both by many laboratories all over the world and from different points of view and with different experimental and theoretic approaches. Among the bacteria, Escherichia coli and Bacillus subtilis serve as model organisms for Gram-negative and Gram-positive organisms, respectively. The huge body of knowledge collected for these organisms calls for an integration of all the data in dedicated databases. We have developed SubtiWiki, a gene and protein-centered database for B. subtilis (1). SubtiWiki not only contains pages for each individual gene and protein, but over the years we have also developed dedicated modules to integrate metabolic pathways, regulation data, and manually curated protein-protein interaction data (2–4). Today, SubtiWiki contains pages for 6013 protein- and RNA-coding genes, information on 1790 operons and links to >6800 publications. Moreover, 49 metabolic and regulatory pathways are presented in intuitive interactive maps. With more than 10 000 page accesses per day, SubtiWiki has become the primary source of information for B. subtilis and other Gram-positive bacteria.
Since the last report on SubtiWiki, we have faced severely impaired accessibility of BsubCyc, the B. subtilis specific database of the BioCyc collection (5). This database has changed its access policy and allows only limited access unless the users acquire a subscription. Thus, features like gene and protein sequences that were previously provided as links to BsubCyc are now directly provided in the new Genome Browser of SubtiWiki. The complexity of annotations collected in SubtiWiki raised a major challenge of data management and called for a more professional system of the database both with respect to the server and the output features. Therefore, we moved away from the MediaWiki engine, and created a more structured database layout and the corresponding server backend. Moreover, we have focused on the development of tools to visualize biological networks, particularly with respect to gene regulation and protein-protein interactions. In addition, studies on gene essentiality and genome minimization have remained in the focus of SubtiWiki. Finally, we have updated the SubtiWiki mobile apps.
THE SubtiWiki ENTRY PAGE AND THE GENE PAGES
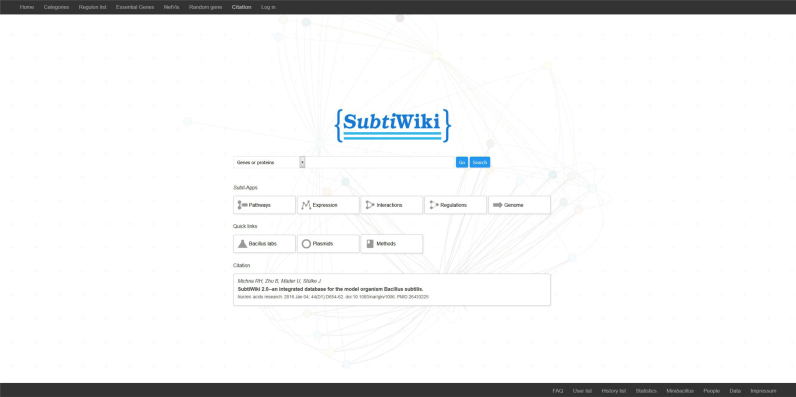
The SubtiWiki entry pages has been re-designed with the aim to provide more easy and direct access to the required set of information (see Figure 1). Novel features of the entry page are direct links to the lists of categories, regulons, and essential genes as well as a direct download link for the simple and intuitive network visualization tool NetVis. Moreover, in the lower bar of the page, the users can directly download all datasets in an Excel-compatible format.
Figure 1.
The entry page of SubtiWiki. The entry page of SubtiWiki provides a simple and clear entry point of the database. The search box guides the user to individual pages of genes/ proteins while the links below provide quick access to the specific browsers and other pages with helpful information.
Even though the underlying structure of the data has been changed from a text-based Wiki to a relational database, the general layout and thus the user experience of the gene and protein pages were only slightly altered. One new feature is the interactive browser for the genomic context. The user can zoom in and out, and scroll up or down the genome using the mouse. Moreover, RNA features can be switched on or off (default status: on). Thus, a wealth of new information on small RNAs that have been discovered in a large transcriptome study is now integrated in SubtiWiki (6,7). By clicking on a gene name in the context browser, the corresponding gene page will open in a new window.
THE GENOME BROWSER
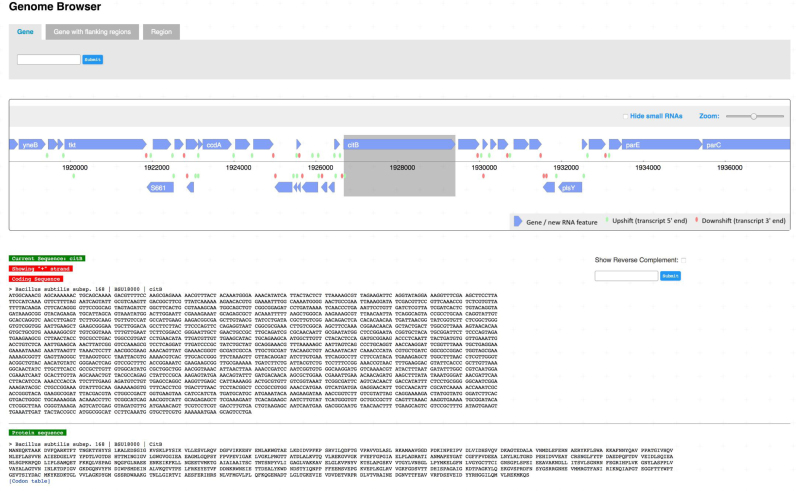
With the newly implemented interactive genome browser, the user can get access to protein and nucleotide sequences (Figure 2). In the browser, the user can scroll along the genome, and by clicking one gene, the corresponding sequences will be shown. In the upper panel, a choice can be made between a particular gene (default), a gene with its flanking sequences (to facilitate primer design or expression signal analysis), or a genomic region. In each case, the selected region will be highlighted by a gray background and the corresponding sequences are shown. Next to the DNA sequence, a search box allows the search for particular sequence motifs (such as restriction sites or protein binding sites) which will then be highlighted in the sequence.
Figure 2.
The genome browser. The genome browser of SubtiWiki provides an integrated view of genomic context as well as the DNA and protein sequences. The user can retrieve a specific gene with or without flanking areas as well as specific regions in the genome. Simple search in the DNA sequence is also enabled.
PRESENTATION OF BIOLOGICAL NETWORKS
Visualization of large scale biological networks has always been an important topic. A proper visualization reveals the underlying structure of a network and enhances its understanding. Therefore, we included visual presentations of protein-protein interaction and regulatory networks in SubtiWiki. The corresponding information has been collected over the years from individual as well as from genome/ proteome level studies. With 5800 and 2400 experimentally validated regulatory and protein-protein interactions in our database, respectively, SubtiWiki has become the prime source of information with respect to gene regulation and protein-protein interactions for B. subtilis (8). In the development of the network browsers, priority was given to the user experience with the aim to have the design of the user interface of the network browsers intuitive and minimal. For this purpose, we selected a gravity model (9), in which nodes are considered as mass points and edges as springs. The global gravity constant is set negative so that nodes repel each other while edges hold them together. The initial layout is circular and positions of nodes are random. Barnes–Hut simulation (10) is used to determine the positions of nodes when all nodes reach force balance.
For better performance, only the subnetwork around a selected gene/protein is displayed in both the interaction and regulation browsers. The initial subnetwork consists of all genes/ proteins that are directly linked to the gene/ protein of interest in the protein–protein interaction or regulatory networks. The regulatory network is treated as undirected network when subnetworks are retrieved. To display further ranging physical or regulatory interactions, a ‘Radius’ can be chosen, which corresponds to the distance level to the selected gene/ protein. The parameter ‘Coverage’ describes the size of the selected subnetwork in comparison to the whole network. It is calculated as the number of genes in the subnetwork over the number of all genes/RNA features and proteins annotated in SubtiWiki for regulatory and protein-protein interaction networks, respectively.
The network browsers (as well as the pathways browser) also allow the users to integrate transcriptomic and proteomic data collected in SubtiWiki. The nodes (genes/ proteins) then are colored with specific color according to expression level and a color legend is shown in the lower left corner.
THE REGULATION BROWSER
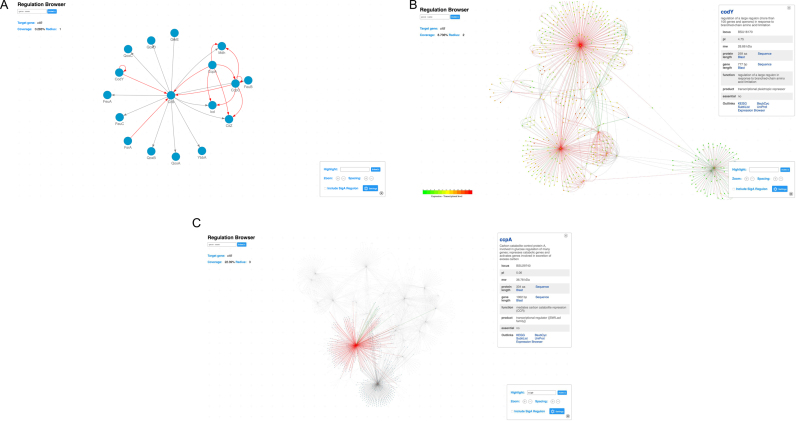
The new regulation browser provides a global view into the regulatory networks in B. subtilis (Figure 3). The search box in the upper left corner allows users to select the gene of interest by name. In the control panel in the lower right corner, the users can change the radius of the subnetwork by zooming in or out (see the three zoom levels for citB gene regulation in Figure 3A–C). The length of the edges can be adjusted by changing the ‘spacing’. The edges in the regulatory network are directed and labelled with a color code according to the regulatory mechanism. We grouped different regulatory mechanisms into three categories, namely positive (e.g. activation, green color), negative (e.g. repression, red color), and other (gray color). The color scheme of the network can be individually adjusted for better contrast. As mentioned above, large scale omics data can be mapped on the regulatory networks (see Figure 3B). An input box allows the users to highlight a series of genes in a large network. When highlighted, those genes stay opaque while the rest of the network fades out (Figure 3C).
Figure 3.
Presentation of regulatory networks using an interactive network browser, using the aconitase-encoding gene, citB, as an example. (A) The initial subnetwork displays the immediate regulators and target genes of citB. (B) With increased radius, more genes are included in this subnetwork. Detailed information of the gene codY is displayed. Nodes in the subnetwork are colored according to its expression level. A legend is displayed at the corner. (C) The subnetwork becomes even larger. At this stage, over 20% of all annotated genes/ proteins/ RNA features are included. The underlying structure of the network is revealed with large interconnecting regulatory clusters clearly displayed. The genes ccpA and sigB are highlighted from the rest of the network.
Expression of the vast majority of genes in B. subtilis depends on the household sigma factor, SigA. Therefore, the SigA regulon is excluded from the subnetworks by default. Ticking the checkbox in the control panel will result in inclusion of this regulon and reload the network. A generic description of a gene will pop up when a node is clicked.
The regulation browser supports the export of the networks. Right click on the blank area brings the menu for different formats. The csv file consists of data for the current subnetwork in adjacency list style. The image is a snapshot of the current viewport. The ‘.nvis’ file is used for the quick network visualization tool NetVis. This format consists of location data of all nodes thus no waiting is needed for stabilization.
NetVis, A SIMPLE NETWORK VISUALIZATION TOOL
SubtiWiki includes an offline tool named NetVis, which applies the same model as in the regulation browser. Users can display networks downloaded from SubtiWiki or visualize their own networks. NetVis supports the individual adjustment of the color scheme, group styling of nodes or edges, and simple manipulation of the network. It requires no installation and is very easy to use.
ESSENTIAL GENES AND MINIMAL GENOMES
Model organisms are central for our understanding of the basic principles of life. In this respect, one set of genes is of particular relevance: The essential genes are absolutely required for the viability of a cell even under optimal conditions. Thus, these genes form the very core of what is required for life. For B. subtilis, the essential gene set has been intensively studied (11–14), and this information has been used to derive a theoretical minimal genome that is based on B. subtilis (15). The experimental verification of this predicted set of 523 and 119 protein- and RNA-coding genes is currently under investigation in the frame of the MiniBacillus project (16). Accordingly, a compilation of all essential genes of B. subtilis is provided in SubtiWiki, and a link to the MiniBacillus project can be found in the lower bar of the entry page.
THE SubtiWiki APP FOR MOBILE DEVICES
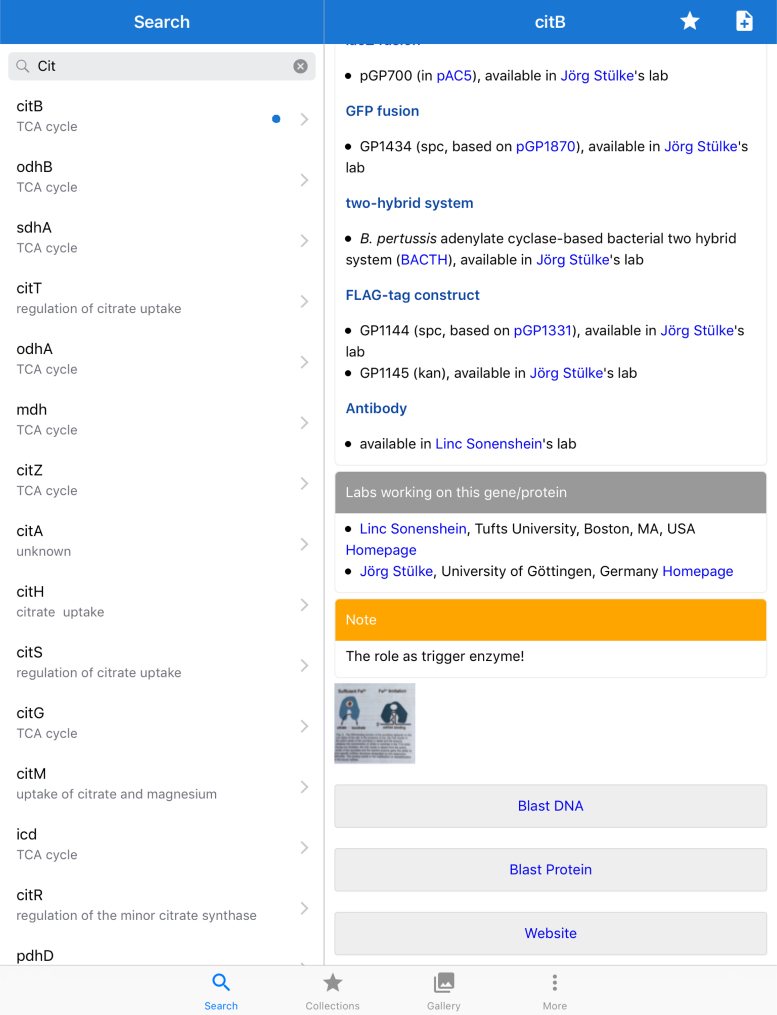
To allow the consultation of SubtiWiki ‘on the go’, we have developed the SubtiWiki app for iOS and Android devices. Since the last presentation, we have directly connected the app to the newly created SubtiWiki relational database. This gives the user direct access to the most recent updates of SubtiWiki. As before, the user can select favourite genes/ proteins and add additional information (text and/ or pictures) to them (Figure 4). This information is strictly private. However, as many scientists use multiple mobile devices, the favourite collections and the user-generated private information are stored on the device and in the iCloud to allow synchronized access on multiple devices of one user. Unfortunately, this feature is not available for Android devices. The SubtiWiki app can be downloaded for free from the Apple App Store and the Google Play Store for iOS and Android users, respectively.
Figure 4.
The SubtiWiki app. The app for mobile devices is currently available in Apple App and Google Play Stores. For iOS users, the new version includes iCloud support for use on multiple devices. The user-generated private notes, images, and favorites are automatically synchronized between the devices.
PERSPECTIVES
SubtiWiki has become one of the most popular databases dedicated to a single organism. Today, it is one of the most complete inventories of knowledge on a living organism in one resource. Importantly, the addition of interactive biological network presentations adds a novel level of usability. We expect, that SubtiWiki can serve as model for the development of databases for many organisms.
ACKNOWLEDGEMENTS
We are grateful to the members of our lab for helpful discussions and encouragement. David Pawlowicz is acknowledged for his help with the development of the interaction browser. We acknowledge the Göttingen Center for Molecular Lifesciences (GZMB) for continued support.
FUNDING
Funding for open access charge: Departmental funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Flórez L.A., Roppel S.F., Schmeisky A.G., Lammers C.R., Stülke J.. A community-curated consensual annotation that is continuously updated: the Bacillus subtilis centred wiki SubtiWiki. Database. 2009; doi:10.1093/database/bap012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lammers C.R., Flórez L.A., Schmeisky A.G., Roppel S.F., Mäder U., Hamoen L., Stülke J.. Connecting parts with processes: SubtiWiki and SubtiPathways integrate gene and pathway annotation for Bacillus subtilis. Microbiology. 2010; 156:849–859. [DOI] [PubMed] [Google Scholar]
- 3. Mäder U., Schmeisky A.G., Flórez L.A., Stülke J.. SubtiWiki – a comprehensive community resource for the model organism Bacillus subtilis. Nucleic Acids Res. 2012; 40:D1278–D1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Michna R., Zhu B., Mäder U., Stülke J.. SubtiWiki 2.0 – an integrated database fort he model organism Bacillus subtilis. Nucleic Acids Res. 2016; 44:D654–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendrasse M., Mueller L.A. et al. . The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/ genome databases. Nucleic Acids Res. 2014; 44:D471–D480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nicolas P., Mäder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., Bidnenko E., Marchadier E., Hoebeke M., Aymerich S. et al. . The condition-dependent whole-transcriptome reveals high-level regulatory architecture in bacteria. Science. 2012; 335:1103–1106. [DOI] [PubMed] [Google Scholar]
- 7. Mars R.A., Nicolas P., Denham E.L., van Dijl J.M.. Regulatory RNA in Bacillus subtilis: a Gram-positive perspective on bacterial NA-mediated gene expression. Microbiol. Mol. Biol. Rev. 2016; 80:1029–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arrieta-Ortiz M.L., Hafemeister C., Bate A.R., Chu T., Greenfield A., Shuster B., Barry S.N., Gallitto M., Liu B., Kacmarczyk T. et al. . An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol. Syst. Biol. 2015; 11:839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fruchterman T.M.J., Reingold E.M.. Graph drawing by force-directed placement. Software – Pract. Exper. 1991; 21:1129–1164. [Google Scholar]
- 10. Barnes J.E., Hut P.. A hierarchical O(n-log-n) force calculation algorithm. Nature. 1986; 324:446–449. [Google Scholar]
- 11. Kobayashi K., Ehrlich S.D., Albertini A., Amati G., Andersen K.K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P. et al. . Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Commichau F.M., Pietack N., Stülke J.. Essential genes in Bacillus subtilis: a re-evaluation after ten years. Mol. Biosyst. 2013; 9:1068–1075. [DOI] [PubMed] [Google Scholar]
- 13. Peters J.M., Colavin A., Shi H., Czarny T.L., Larson M.H., Wong S., Hawkins J.S., Lu C.H.S., Koo B.M., Marta E. et al. . A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell. 2016; 165:1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koo B.M., Kritikos G., Farelli J.D., Todor H., Tong K., Kimsey H., Wapinski I., Galardini M., Cabal A., Peters J.M. et al. . Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 2017; 4:291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reuß D.R., Commichau F.M., Gundlach J., Zhu B., Stülke J.. The blueprint of a minimal cell: MiniBacillus. Microbiol. Mol. Biol. Rev. 2016; 80:955–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reuß D.R., Altenbuchner J., Mäder U., Rath H., Ischebeck T., Sappa P.K., Thürmer A., Guérin C., Nicolas P., Steil L. et al. . Large-scale reduction of the Bacillus subtilis genome: consequences for the transcriptional network, resource allocation, and metabolism. Genome Res. 2017; 27:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]