Abstract
MINTbase is a repository that comprises nuclear and mitochondrial tRNA-derived fragments (‘tRFs’) found in multiple human tissues. The original version of MINTbase comprised tRFs obtained from 768 transcriptomic datasets. We used our deterministic and exhaustive tRF mining pipeline to process all of The Cancer Genome Atlas datasets (TCGA). We identified 23 413 tRFs with abundance of ≥ 1.0 reads-per-million (RPM). To facilitate further studies of tRFs by the community, we just released version 2.0 of MINTbase that contains information about 26 531 distinct human tRFs from 11 719 human datasets as of October 2017. Key new elements include: the ability to filter tRFs on-the-fly by minimum abundance thresholding; the ability to filter tRFs by tissue keywords; easy access to information about a tRF’s maximum abundance and the datasets that contain it; the ability to generate relative abundance plots for tRFs across cancer types and convert them into embeddable figures; MODOMICS information about modifications of the parental tRNA, etc. Version 2.0 of MINTbase contains 15x more datasets and nearly 4x more distinct tRFs than the original version, yet continues to offer fast, interactive access to its contents. Version 2.0 is available freely at http://cm.jefferson.edu/MINTbase/.
INTRODUCTION
Compared to microRNAs (miRNAs), tRNA-derived fragments (tRFs) are more diverse and can arise from either the precursor or the mature tRNA. While the tRF field is relatively new, significant progress has been made to elucidate their types and functions. This is evidenced by an increasing number of publications by others and us providing strong evidence that tRFs are an important new category of regulatory molecules (1–20). Several recent reviews offer comprehensive summaries of what is currently known about these molecules (18,21–23). In addition to human, tRFs have been reported in many other organisms, including mouse (3), fruit-fly (10,24), plants (4) and prokaryotes (25).
Our previous analyses (8,17,20,26,27) and the work we present below focus on tRFs that overlap the mature tRNAs. Structurally, these tRFs fall into five distinct classes (20,27): 5′-tRNA halves, 3′-tRNA halves, 5′-tRFs, 3′-tRFs and the newly discovered, rich category of ‘internal tRFs’ (i-tRFs). We presented and discussed the characteristics of the i-tRF class previously (20). There are also tRFs, known as tsRNAs and 3′-U tRFs, that originate from cleavage of the precursor tRNA molecule (28–30). Recently, tsRNAs were shown to be dysregulated in human cancers (28) and to give rise to cancer-specific signatures (31). Because tsRNAs are derived from the precursor tRNA molecule, they require a different type of analytical work than fragments that overlap with the mature tRNA, and as such, we have slated them for inclusion in an upcoming release of MINTbase.
For tRFs overlapping the mature tRNA, several mechanisms of tRF action have been established by now. For example, some tRFs have been shown to be loaded on Argonaute and, thus, to act like miRNAs—see (BioRxiv: https://doi.org/10.1101/143974) and (3,5,20). Other tRFs have been shown to compete with mRNAs for the binding to RNA-binding proteins (6). tRNA halves have also been observed in regulatory and direct physical interactions with several proteins and protein complexes that include cytochrome C (32), ribosomes (33) and the multi-synthetase complex (11). Moreover, tRFs can also act as piRNAs (8).
In recent work, we reported that tRFs are produced constitutively in human cells, in health and disease, and that their composition and abundances are shaped by an individual’s sex, population origin, race, tissue and disease subtype (20). These properties of tRFs are in complete analogy to what we reported previously for the category of short ncRNAs known as miRNA isoforms or ‘isomiRs’ (34,35). These similarities prompted us to extend our original analyzes of 768 human datasets to the >11 000 short RNA-seq datasets from The Cancer Genome Atlas (TCGA).
MATERIALS AND METHODS
Deterministic and exhaustive mining of tRFs
A necessary requirement prior to embarking on our studies was the development of a specialized mapper for sequenced reads. The mapper needed to be deterministic, exhaustive and to take into account the repeat nature of tRNAs and the idiosyncrasies of the human genome architecture. One key advantage of MINTmap is that, for datasets that are not in colorspace, it can carry out the mining of tRFs without any need to map the sequenced reads on the genome while guaranteeing that the mining is deterministic and exhaustive. For colorspace inputs, we describe the needed preliminary steps in (27). The MINTmap codes are freely available at https://cm.jefferson.edu/MINTcodes/. For the analysis of the TCGA datasets, we used the default parameter settings of MINTmap.
Thresholding of the datasets
We mined each TCGA dataset using our recently developed MINTmap algorithm (27). We retained those of the identified tRFs that exceeded a normalized abundance of 1 RPM and entered them into MINTbase. Users can use the newly-provided on-the-fly filtering capability to sub-select among tRFs from TCGA and from the datasets of v1.0 based on their abundance.
TCGA datasets and tRF profiling
We downloaded the TCGA datasets on 16 October 2015 from TCGA’s Cancer Genomic Hub. We used MINTmap to process separately each of 11 198 short RNA-seq datasets. The union of tRFs that survived the mining and filtering process were entered into MINTbase. It is important to note here a considerable limitation of the short RNA-seq TCGA datasets that affects the enumeration of tRNA halves. Specifically, the TCGA datasets were generated by running deep-sequencing PCR for 30 cycles. Recall here that, by definition, tRNA halves are longer than 30 nt. Consequently, tRNA halves are not represented among the TCGA datasets. Even with this limitation, the TCGA module of MINTbase is a treasure trove of invaluable tRF information because tRNA halves represent a small fraction of all tRFs that are known to overlap the mature tRNAs.
Data visualization
The data visualizations appearing on the ‘summary vista’ of MINTbase were made using the Highcharts JavaScript library (http://highcharts.com).
RESULTS
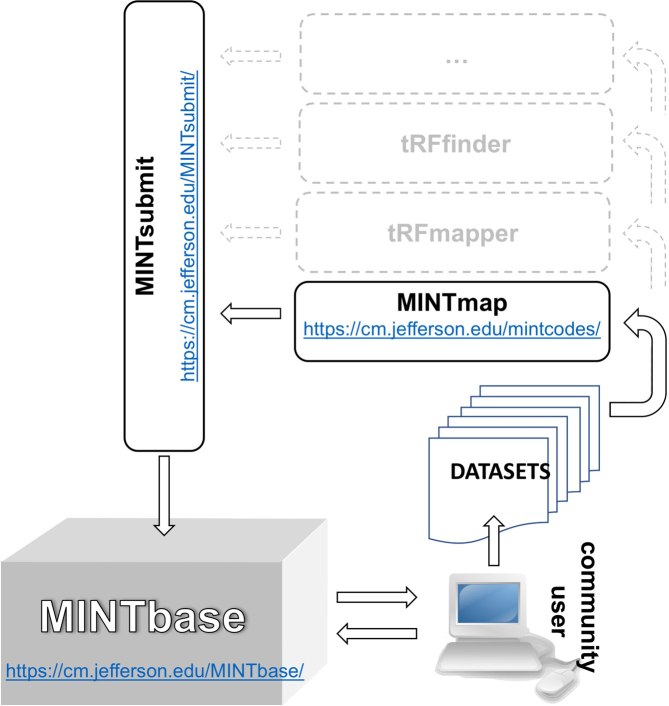
In this section, we highlight several new capabilities that we implemented and incorporated in version 2.0 of MINTbase. MINTbase is web-accessible, has been populated with nuclear and mitochondrial (MT) tRFs from human tissues, and is the central element of a solution that we designed for the study of these molecules (Figure 1). MINTbase can accommodate tRFs that have been mined using any of the available tools such as MINTmap (27), tDRmapper (36), tRFfinder (37), etc.
Figure 1.
MINTbase, MINTsubmit and the flow of information. We used the MINTmap algorithm to mine nearly 12 000 public datasets for tRFs that we have incorporated in version 2.0 of MINTbase. Users can interact directly with MINTbase. Users can also process their own datasets using MINTmap or other tRF mining tools, such as tDRmapper, tRFfinder, etc. and optionally contribute their findings for inclusion in MINTbase.
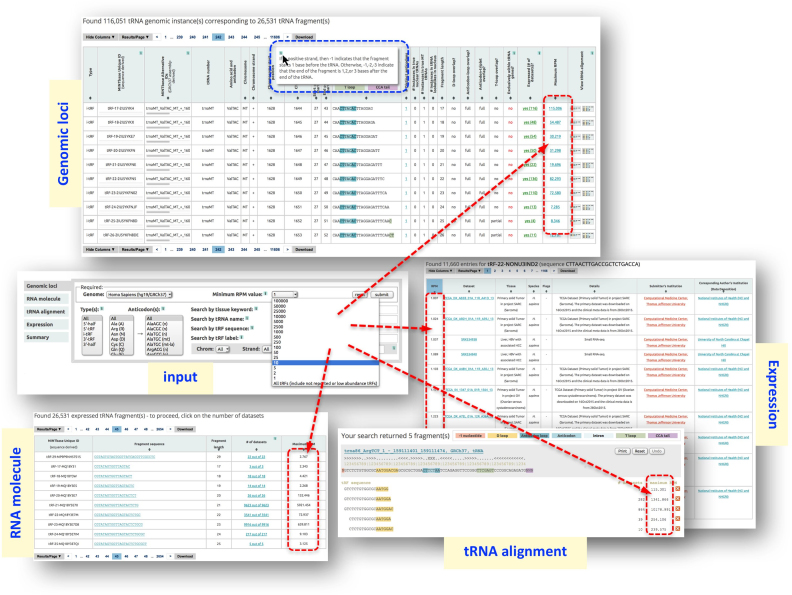
MINTbase consists of five ‘vistas’, each offering a different vantage point from which to study tRFs: genomic loci, RNA molecule, tRNA alignment, expression, and summary. The ‘genomic loci’ vista focuses on the genomic instances of each tRF, is the most detailed vista of MINTbase, and is genome-centric. The ‘RNA molecule’ vista is molecule-centric and offers basic information for the tRFs. The ‘summary’ vista provides access to all available information about a tRF or a full-length mature tRNA, including graphs of its expression profile across various dataset collections. Each possible tRF sequence with lengths between 16 and 50 nt already has its unique summary record in MINTbase, independently of whether there is evidence of transcription. The ‘expression’ vista provides a portal to the literature and the datasets containing a given tRF, including information about the tissue in which the tRF is expressed, its abundance, the PubMed identifier of the publication that first described the dataset from which the tRF was mined, etc. Finally, the ‘tRNA alignment’ vista is mature-tRNA-centric (at the isodecoder level) and presents an alignment of the expressed tRFs that map to the isodecoder at hand together with their maximum abundances, and the number of datasets that contain each of the shown tRFs at the user-selected abundance threshold.
Filtering database searches by abundance
An important feature that we introduce in version 2.0 is the ability to filter tRFs based on their normalized abundance. The normalization used is reported in RPM. As we analyzed more and more datasets, it became evident that a tRF’s abundance varies characteristically across tissues or different diseases of the same tissue, while remaining essentially unchanged within samples of the same type (20). Thus, we viewed the ability to filter user searches by RPM value as a sine qua non feature of version 2.0 of MINTbase. The user-selectable minimum abundance thresholds range from 1 RPM (default) to 100 000 RPM (Figure 2). Imposing a minimum abundance constraint can be selected ‘on-the-fly’ and combined with other criteria during a search.
Figure 2.
In version 2.0, we incorporated tRF abundance information for all mined tRFs and across all analyzed datasets. The abundance constraints are relevant for and thus available in four of the five available vistas.
Filtering database searches by keywords (misspellings allowed!)
Another key feature that we introduced in version 2.0 is the ability to restrict the searches by using keywords. Keywords are compared to the description of the analyzed datasets. For example, the user can search for tRFs only in datasets originating from ‘breast’ tissue, only in datasets that are part of the lung adenocarcinoma (LUAD) subset of the TCGA repository, etc. The allowed keywords are flexible and can include partial words (e.g. ‘live’ instead of liver), or misspelled words (e.g. ‘lever’ instead of liver). The user can leave the keyword filter empty to retrieve information from all datasets in MINTbase. The ability to filter by keywords is available to all vistas except the summary vista.
Flexibility in using tRNA labels
In the absence of an accepted standard, several tRF labeling schemes have been proposed and are currently in use in the literature. With that in mind, we made MINTbase capable of handling user searches that can be issued using any of several tRNA labels. The allowed labels include: gtRNAdb IDs (38), HUGO Gene Nomenclature Committee (HGNC) symbols, legacy IDs (39), genome-centric tRNA labels (20) and the genome-assembly independent tRF license plates (40).
We introduced the last two labeling schemes in recent work, in an effort to address two specific requirements. The genome-centric tRNA labels (20) are meant to distinguish among isodecoders that share sequence segments yet are located at different parts of the genome; also, they are meant to specifically indicate the location within the mature tRNA where a tRF is located. On the other hand, the tRF license plates (40) can be deduced from the tRF sequence alone, and are compact, unique and genome-assembly independent. Because, a tRF’s license plate is universally unique, ‘minting’ it does not require a centralized brokering mechanism that issues identifiers. Anyone can generate the license plate of his/her favorite tRF or deduce the tRF from a license plate, by using the free and portable codes we make available at https://cm.jefferson.edu/MINTcodes/. Thus, researchers working in parallel with the same potentially unpublished tRF at different institutions will generate and use the same exact label for it, which will greatly facilitate subsequent cross-checking and consistency of nomenclature.
New form style, flexibility and connectivity across vistas
Version 2.0 features a new layout and form style that makes it easier to distinguish between vistas. If a search produces multiple results that cannot be presented simultaneously in the selected vista, a ‘disambiguation’ page is generated automatically and prompts the user to select among the possible answers before proceeding. In another important change, we increased the cross-links to enable swift navigation between the vistas.
Context-sensitive help
To better deliver information during a user’s interaction with MINTbase, we expanded the presence of context-sensitive help. In version 2.0, every page of MINTbase, whether an input or an output page, contains multiple information icons that provide brief, context sensitive summaries. More detailed information is available on the help pages of MINTbase.
How to combine keywords and abundance-constraints
As an example, let us say that the user wishes to explore the abundant tRFs in the ovarian cancer (OV) datasets of TCGA. The user can select the ‘RNA molecule’ vista, type ‘TCGA_OV’ in the ‘Search by tissue keyword’ window and restrict the ‘minimum RPM value’ to e.g. 100 using the pull-down menu in order to focus on abundant tRFs. A total of 388 tRFs satisfy these criteria as of October 2017. The contents of the output page can be sorted by a number of user-selectable criteria, such as decreasing maximum abundance (Figure 3A). A striking tRF among those retrieved by this search is CTTAACTTGACCGCTCTGACCA, a 3′-tRF from the MT tRNAValTAC. This tRF is present in most (418 of the 499) TCGA_OV datasets at an abundance ≥ 100 RPM, and reaches a maximum RPM value of 35 972. The last column of CTTAACTTGACCGCTCTGACCA’s row links to a rendering of its alignment to the parental tRNA isodecoder (Figure 3B).
Figure 3.
Panel (A) shows the output of a query that is constrained by the keyword ‘TCGA_OV’ and by the minimum RPM value of ‘100’. The 388 tRFs that satisfy the user’s criteria are shown sorted in order of decreasing maximum abundance—see also text. Panel (B) shows the most abundant 3′-tRF aligned to the corresponding parental tRNA template. This tRF is present in 418 of the 499 TCGA_OV datasets. Clicking on the ‘print’ button allows the user to generate a PDF file with the shown alignment.
New and diverse information at the user’s fingertips
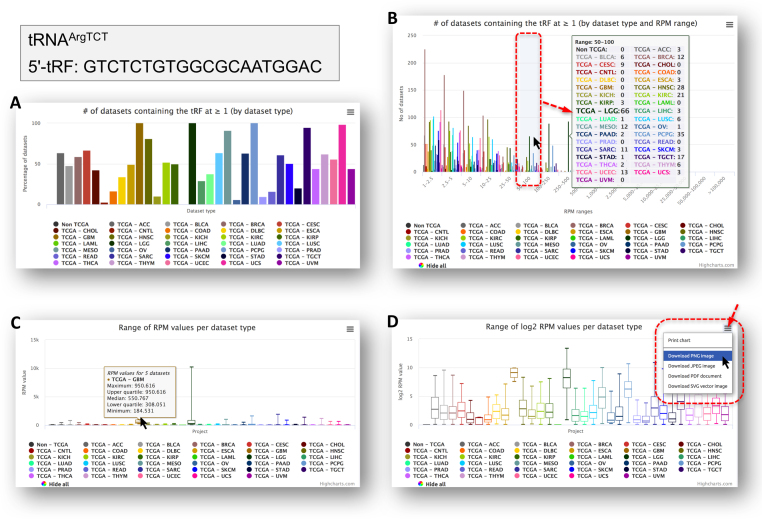
To highlight the new information that is available in version 2.0 of MINTbase we focus on the summary vista and on GTCTCTGTGGCGCAATGGAC, which is a 5′-tRF from tRNA-Arg-TCT-4-1. Figure 4A shows what portion of the datasets contain this tRF, across each of the various categories (32 cancer types plus non-cancer datasets). Note how, for four cancers, kidney chromophobe (KICH), acute myeloid leukemia (LAML), OV, and prostate adenocarcinoma (PRAD), this tRF is present in fewer than 10% of the corresponding datasets. Figure 4B shows the distribution (Y-axis) of the tRF’s RPM abundance (X-axis) across the datasets in which it is present. The next two panels of Figure 4 show the range of RPM values for this tRF in linear (C) and in logarithmic (D) scales. It is evident that this tRF is highly expressed in a considerable portion of the lower grade glioma (LGG) datasets. Popovers at each of the bars in these charts provide more detailed information. Lastly, the user can select which cancer types to visualize by clicking on the cancer labels below each chart. Each chart can be printed or exported to a variety of formats (Figure 4D).
Figure 4.
A sampling of the diverse information that is available for each tRF in version 2.0 of MINTbase. Panels (A) through (D) provide different aspects of the available information for the 5′-tRF GTCTCTGTGGCGCAATGGAC from tRNAArgTCT. See text for more information.
Augmented tRF records and inclusion of MODOMICS information
The finite number of human nuclear and MT tRNAs means that we can enumerate the space of all possible tRFs that overlap with mature tRNAs and have a given range of lengths. Currently, there are 594 972 such sequences with lengths between 16 and 50 nt. Thus, we created a summary record for each of these possible tRFs. In version 2.0, for those tRFs with abundance ≥1.0 RPM in a public dataset, we augmented each record by including graphs that show the tRF’s expression patterns and abundances across the datasets of MINTbase. For the TCGA datasets, we provide additional granularity by showing this information separately for each of 32 TCGA cancer types. All of the graphs are interactive and provide options for ‘downloading’ and ‘printing.’ Another addition in version 2.0 is the inclusion of summary records for each full-length parental mature tRNAs that list the known RNA modifications contained in the Modomics database (41). The summary records also include direct links to the corresponding Modomics web pages. Additionally, these records contain graphs that show where in the mature tRNA the tRFs begin and end, and the number of tRFs in each category that are produced from this tRNA.
Helping make new discoveries: an example
We will use the newly reported category of i-tRFs to demonstrate how one can mine MINTbase to generate new findings. Because i-tRFs were reported for the first time only recently (20), there is limited information in the literature about their attributes across human tissues. For example: How many i-tRFs exist? How many are of ambiguous genomic origin? How many are abundantly expressed across human tissues? Are they produced by nuclear tRNAs? MT tRNAs? Or both? Are they produced preferentially by some tRNAs, and if so, which ones? MINTbase can help answer these questions easily. In the ‘RNA molecule’ vista, the user selects ‘i-tRFs’ in the ‘Type(s)’ column leaving the amino acid and isoacceptor settings to their default values (‘All’). To restrict the search to only abundant i-tRFs, the user can further set the ‘minimum RPM value’ to 100. The search returns 1 088 distinct molecules that can be found in any of 6 618 distinct genomic locations as of October 2017. The output page can be mirrored locally by clicking on the ‘Download’ button adjacent to the Table and imported into Excel. Minimal processing in Excel reveals that 541 of these i-tRFs originate exclusively in tRNA space and have lengths largely between 18 and 25 nt. For 94 of these 541 i-tRFs the parental isodecoder can be defined unambiguously (column 9 contains the value ‘1’). Of these 94 i-tRFs, 69 or 73.4% originate in MT tRNAs (column 6), primarily in tRNAHisGTG(mt) (19 i-tRFs), tRNALeuTAG(mt) (12 i-tRFs) and tRNASerGCT(mt) (11 i-tRFs).
Comparison with other databases
Here, we summarize the key elements distinguishing version 2.0 of MINTbase from tRFdb (42) and tRF2Cancer (37). A more detailed comparison can be found in (27).
tRFdb is powered by the BLAST search tool (43). BLAST is probabilistic and, thus, cannot guarantee an exhaustive enumeration of all the genomic hits for a read. tRF2Cancer relies on the tool tRFfinder that was developed by the same authors and assumes that reads are randomly distributed across the length of the tRNA. This assumption is not supported by the available public data and leads, we believe, to an over-representation of tRFs arising from the 5′ or the 3′ end of the tRNA. On the other hand, MINTbase relies on the deterministic and exhaustive MINTmap algorithm that outperforms tRFdb and tRF2Cancer in both sensitivity and specificity (27). This is particularly evident when comparing MINTbase with tRF2Cancer: MINTbase includes 23 413 tRFs with abundance ≥1.0 RPM that were mined from TCGA whereas tRF2Cancer lists only 606.
Another important difference pertains to the size of fragments that are included in these databases. For example, both tRFdb and tRF2Cancer include fragments as short as 14 nt. The problem with such short sequences is that they are more likely to arise from non-tRNA locations of the genome (26). The same can happen with longer sequences as well, and MINTbase tackles this by (i) considering only fragments that are ≥16 nt, and, (ii) explicitly reporting whether a given fragment is exclusive to tRNA space or can be found elsewhere on the genome. Doing so alerts the user to the possibility that some tRFs could be false-positives.
Three more elements of MINTbase version 2.0 differentiate it from tRFdb and tRF2Cancer: (i) the inclusion of MT tRFs; (ii) the inclusion of the new category of i-tRFs; and (iii) the ability to filter searches using tRF abundances. These elements are missing currently from tRF2Cancer. tRFdb includes a few i-tRFs albeit not explicitly and no MT tRFs. A quick search shows that TCGA comprises 3 063 MT tRFs and 6 625 i-tRFs, each with an abundance ≥ 5.0 RPM in at least one TCGA dataset. Recall here that i-tRFs and MT-tRFs are not mutually exclusive categories (i-tRFs can arise from both nuclear and MT tRNAs). Moreover, version 2.0 of MINTbase also provides advanced search capabilities, as we described above.
At last, we want to stress an important design element that differentiates MINTbase from the other databases. Specifically, MINTbase is meant to be a ‘living’ repository that easily accepts contributions from the research community. We describe this next.
Going beyond TCGA—MINTsubmit enables contribution of datasets by the community
MINTbase derives its value from the public datasets that are contained in it. We designed the database so that it can translate the contributions of individual teams into a resource that can benefit the entire research community with minimal intervention on our part.
The initial release of MINTbase (version 1.0) included tRFs from 768 human datasets. Version 2.0 demonstrates what can be gained from increasing the number and diversity of datasets in MINTbase. The much larger repository, combined with version 2.0’s enhanced capabilities, permits elaborate comparisons across tissues, conditions, isodecoders, isoacceptors, etc. The tool MINTsubmit (37) facilitates the incorporation of new datasets in MINTbase. Its user-friendly interface allows scientists who have generated tRFs in their own work to submit them for inclusion in MINTbase. MINTsubmit automatically verifies the identity of the submitter, and ensures that credit is given to the team that generated the data by permanently linking the team’s name to the respective MINTbase entries. To ensure fairness, MINTsubmit only accepts data that (i) already exist in a public repository (e.g. NIH Gene Expression Omnibus or GEO) and (ii) are described in an article already on PubMed. New datasets typically appear on MINTbase a few days following submission.
DISCUSSION
We presented the key new elements of version 2.0 of MINTbase, a database of human nuclear and MT tRFs. Version 2.0 comprises information that we culled by analyzing an additional 11 198 public datasets from TCGA. Compared to the previous version, version 2.0 represents a ≥15x increase in the number of datasets. Also, version 2.0 contains information on 26 531 tRFs, a nearly 4x increase over version 1.0.
During the implementation of version 2.0, we constrained ourselves by imposing two key design criteria: speed and search flexibility. To achieve speed, we made judicious use of mysql databases that we pre-populated with the output of MINTmap from our mining of RNA-seq datasets. A layer of relational tables enabled complex queries from within MINTbase’s five ‘vistas.’ Currently, complicated searches as well as searches that retrieve many sequences from the database, complete in 1–2 s (wall-clock time).
Version 2.0 of MINTbase provides users with access to a much larger collection of tRFs that are cross-linked in various combinations and can be connected back to the nearly 12 000 public datasets contained in MINTbase. As more researchers embark on studies of these important molecules, we expect more, unique and novel tRFs from human tissues and across different diseases and conditions to be identified and become publicly available. As more members of the community use MINTsubmit, we also expect the contributions to MINTbase to increase. In addition, as knowledge accumulates and the molecular mechanisms of tRF actions become clear, we anticipate including new modules in MINTbase, like target predictions. In the meantime, we hope that MINTbase and MINTmap (27) will prove useful additions to the toolbox of the research groups that study these molecules.
ACKNOWLEDGEMENTS
Authors’ Contributions: I.R. oversaw the development of version 2.0 of MINTbase. V.P., P.L., I.R., A.G.T., E.L., M.S., Y.K. and R.M. designed various aspects of version 2.0 of MINTbase. V.P. and P.L. implemented version 2.0 of MINTbase. P.L. mined all TCGA datasets for tRFs using MINTmap. R.M. analyzed data. I.R., A.G.T., P.L., V.P., R.M., E.L., M.S. and Y.K. extensively tested version 2.0 under multiple settings prior to release. I.R., V.P., P.L. and A.G.T. wrote the manuscript. All authors have read and approved the final manuscript.
FUNDING
William Keck Foundation Grant (to IR) (in part); NIH/NCI R21-CA195204 (IR) (in part); Institutional Funds. Funding for open access charge: Institutional Funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Anderson P., Ivanov P.. tRNA fragments in human health and disease. FEBS Lett. 2014; 588:4297–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blanco S., Dietmann S., Flores J.V., Hussain S., Kutter C., Humphreys P., Lukk M., Lombard P., Treps L., Popis M. et al. . Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014; 33:2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kumar P., Anaya J., Mudunuri S.B., Dutta A.. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014; 12:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Loss-Morais G., Waterhouse P.M., Margis R.. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol. Direct. 2013; 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R.. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goodarzi H., Liu X., Nguyen H.C., Zhang S., Fish L., Tavazoie S.F.. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015; 161:790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goodarzi H., Nguyen H.C.B., Zhang S., Dill B.D., Molina H., Tavazoie S.F.. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell. 2016; 165:1416–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Honda S., Kawamura T., Loher P., Morichika K., Rigoutsos I., Kirino Y.. The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res. 2017; 45:9108–9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Honda S., Loher P., Shigematsu M., Palazzo J.P., Suzuki R., Imoto I., Rigoutsos I., Kirino Y.. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3816–E3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karaiskos S., Grigoriev A.. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000 Res. 2016; 5:2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keam S.P., Sobala A., Ten Have S., Hutvagner G.. tRNA-derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J. Proteome Res. 2017; 16:413–420. [DOI] [PubMed] [Google Scholar]
- 12. Lowe T.M., Chan P.P.. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016; 44:W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mami I., Pallet N.. Transfer RNA fragmentation and protein translation dynamics in the course of kidney injury. RNA Biol. 2015; doi:10.1080/15476286.2015.1107704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olvedy M., Scaravilli M., Hoogstrate Y., Visakorpi T., Jenster G., Martens-Uzunova E.. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. 2016; 7:24766–24777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schifano J.M., Cruz J.W., Vvedenskaya I.O., Edifor R., Ouyang M., Husson R.N., Nickels B.E., Woychik N.A.. tRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res. 2016; 44:1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schorn A.J., Gutbrod M.J., LeBlanc C., Martienssen R.. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017; 170:61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shigematsu M., Kirino Y.. 5΄-Terminal nucleotide variations in human cytoplasmic tRNAHisGUG and its 5΄-halves. RNA. 2017; 23:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shigematsu M., Kirino Y.. tRNA-derived short non-coding RNA as interacting partners of argonaute proteins. Gene Regul. Syst. Bio. 2015; 9:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Telonis A.G., Kirino Y., Rigoutsos I.. Mitochondrial tRNA-lookalikes in nuclear chromosomes: could they be functional. RNA Biol. 2015; 12:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Telonis A.G., Loher P., Honda S., Jing Y., Palazzo J., Kirino Y., Rigoutsos I.. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015; 6:24797–24822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keam S.P., Hutvágner G.. tRNA-Derived Fragments (tRFs): Emerging New Roles for an Ancient RNA in the Regulation of Gene Expression. Life. 2015; 5:1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar P., Kuscu C., Dutta A.. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci. 2016; 41:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ibba M. Transfer RNA comes of age. RNA. 2015; 21:648–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grigoriev A., Bonini N.M.. Age-dependent patterns of microRNA RISC loading. Aging. 2014; 6:705–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gebetsberger J., Polacek N.. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013; 10:1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Telonis A.G., Loher P., Kirino Y., Rigoutsos I.. Consequential considerations when mapping tRNA fragments. BMC Bioinformatics. 2016; 17:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loher P., Telonis A.G., Rigoutsos I.. MINTmap: fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci. Rep. 2017; 7:41184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pekarsky Y., Balatti V., Palamarchuk A., Rizzotto L., Veneziano D., Nigita G., Rassenti L.Z., Pass H.I., Kipps T.J., Liu C.G. et al. . Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:5071–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A.Z., Kay M.A.. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010; 16:673–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee S.R., Collins K.. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005; 280:42744–42749. [DOI] [PubMed] [Google Scholar]
- 31. Balatti V., Nigita G., Veneziano D., Drusco A., Stein G.S., Messier T.L., Farina N.H., Lian J.B., Tomasello L., Liu C.G. et al. . tsRNA signatures in cancer. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:8071–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saikia M., Jobava R., Parisien M., Putnam A., Krokowski D., Gao X.H., Guan B.J., Yuan Y., Jankowsky E., Feng Z. et al. . Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell Biol. 2014; 34:2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P.. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011; 43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loher P., Londin E.R., Rigoutsos I.. IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies. Oncotarget. 2014; 5:8790–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Telonis A.G., Loher P., Jing Y., Londin E., Rigoutsos I.. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015; 43:9158–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Selitsky S.R., Sethupathy P.. tDRmapper: challenges and solutions to mapping, naming and quantifying tRNA-derived RNAs from human small RNA-sequencing data. BMC Bioinformatics. 2015; 16:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng L.L., Xu W.L., Liu S., Sun W.J., Li J.H., Wu J., Yang J.H., Qu L.H.. tRF2Cancer: a web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res. 2016; 44:W185–W193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan P.P., Lowe T.M.. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016; 44:D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan P.P., Lowe T.M.. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009; 37:D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pliatsika V., Loher P., Telonis A.G., Rigoutsos I.. MINTbase: a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics. 2016; 32:2481–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al. . MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumar P., Mudunuri S.B., Anaya J., Dutta A.. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015; 43:D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]