Abstract
The Encyclopedia of DNA Elements (ENCODE) Data Coordinating Center has developed the ENCODE Portal database and website as the source for the data and metadata generated by the ENCODE Consortium. Two principles have motivated the design. First, experimental protocols, analytical procedures and the data themselves should be made publicly accessible through a coherent, web-based search and download interface. Second, the same interface should serve carefully curated metadata that record the provenance of the data and justify its interpretation in biological terms. Since its initial release in 2013 and in response to recommendations from consortium members and the wider community of scientists who use the Portal to access ENCODE data, the Portal has been regularly updated to better reflect these design principles. Here we report on these updates, including results from new experiments, uniformly-processed data from other projects, new visualization tools and more comprehensive metadata to describe experiments and analyses. Additionally, the Portal is now home to meta(data) from related projects including Genomics of Gene Regulation, Roadmap Epigenome Project, Model organism ENCODE (modENCODE) and modERN. The Portal now makes available over 13000 datasets and their accompanying metadata and can be accessed at: https://www.encodeproject.org/.
INTRODUCTION
The sequencing and annotation of the human genome is an ongoing community effort and is made possible by the collective approaches of several large consortia and individuals. In 2003, the NHGRI formed the Encyclopedia of DNA Elements (ENCODE) project with the aim of bringing together a variety of experimental and computational labs to identify biochemically active regions in the human and mouse genomes utilizing a variety of high-throughput approaches (1). This corpus of data provides an astounding resource for annotation, curation and functional characterization in the human and mouse genomes in a large variety of sample types (2,3).
The ENCODE Data Coordinating Center (DCC) is a centralized resource composed of individuals that possess the technical and scientific expertise to work with scientists, computational analysts and engineers. We work collaboratively to bring transparency in the methods used to promote data dissemination, interoperability, standardization and reproducibility. In concert with the submitting labs, we perform substantial curation and vetting of the data deposited into the DCC leading to uniform reporting of (meta)data metrics, across labs and as a function of time.
To date, ENCODE alone has produced over 9000 high-throughput sequencing libraries from assays such as: RNA-Seq, chromatin immunoprecipitation (ChIP)-Seq (Transcription Factors and Histone Modifications), ATAC-Seq, DNase-Seq and assays designed to capture chromosomal conformations (ChIA-PET and Hi-C) (4). The flexibility inherent in these assays and sequencing platforms allows for the rapid development of protocols and optimizations but creates challenges with respect to accurately tracking and reporting metadata to discern important and distinct features underlying the creation of each dataset.
Although the ENCODE Portal was designed to accommodate the data from the ENCODE project, it has become a continually evolving resource and now includes metadata and data from related genomics projects like The Roadmap Epigenomes Project (REMC, (5,6)) and modENCODE (7–9). We are also the primary DCC for the modERN and Genomics of Gene Regulation (GGR, https://www.genome.gov/27561317/genomics-of-gene-regulation/) projects.
THE PORTAL
New data at the ENCODE Portal
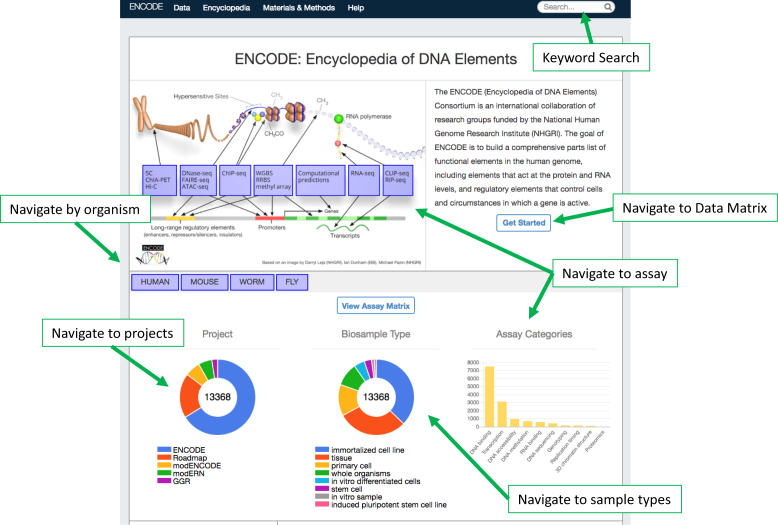
To date, there are over 13000 datasets available through the Portal from human, mouse, Drosophila and Caenorhabditis elegans assayed under a variety of different physiological conditions, (Figure 1), totaling over 500 terabytes of data. The Portal landing page has an interactive user interface that illustrates the breakdown of the data according to project, organism, biological sample type and assay for quick access. As a genomic data resource, we provide persistent identifiers that can be used to access even revoked or archived data.
Figure 1.
The Portal Landing Page. The Portal landing page is arranged to make it easy to navigate and dig into particular data types. In addition to the drop-down menu bar on top, there is a keyword search function—through which genes, cell lines and other ontological terms can be queried and datasets scored with those properties retrieved. Clickable bars allow the user to navigate to data derived from a particular organism, sample type, project and/or assay. A quick link to the Data Matrix is also available.
New Portal views
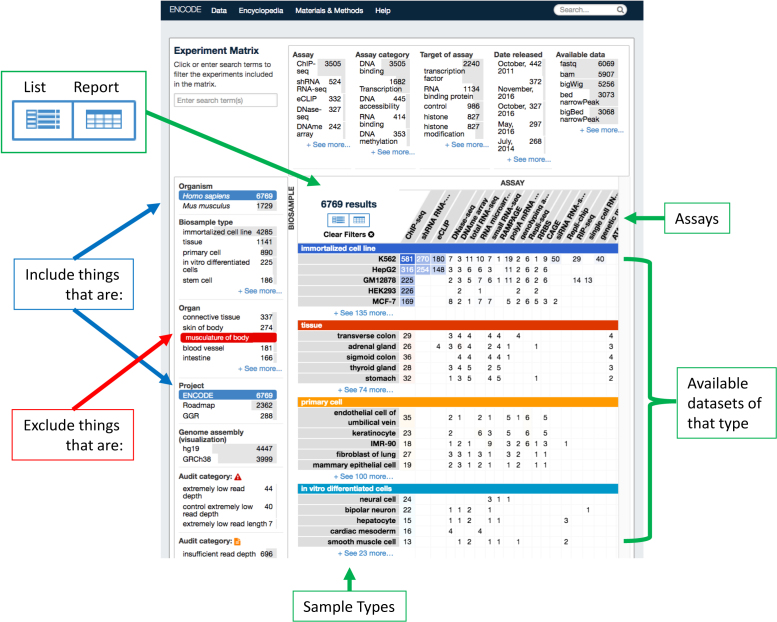
To make finding data easier, we have expanded the summary views from a simple list view to a data matrix and report table. The data matrix condenses the experiments into an array of sample types versus assays conducted on a given sample type, (Figure 2). The faceted browsing interface on the left and top let you select for (blue highlight) and against (red highlight) different features of the data throughout all the available views. The results appear in the matrix on the right and indicate the number of experiments that fall within the chosen set of parameters. Lists of individual experiments can then be accessed by navigating the matrix and clicking on desired cells or by clicking on the list icon in the upper left-hand corner of the matrix view (Figure 2). Collectively, this allows the end-user to get a high-level overview of the data structure.
Figure 2.
The Data Matrix. Screenshot of the Data Matrix. To the left of the matrix are faceted search bars that can be used to positively selected (blue) and negatively select (red) for certain data types on several metadata properties. The matrix itself correspond to a count (with links) to datasets done on certain sample types by certain assays. Clicking on the count—takes the user to a list of the datasets, as does clicking on the ‘list’ icon. Metadata for the data displayed in the Data Matrix and be obtained by clicking on the ‘report’ icon and downloaded in .tsv format.
Metadata structure and download
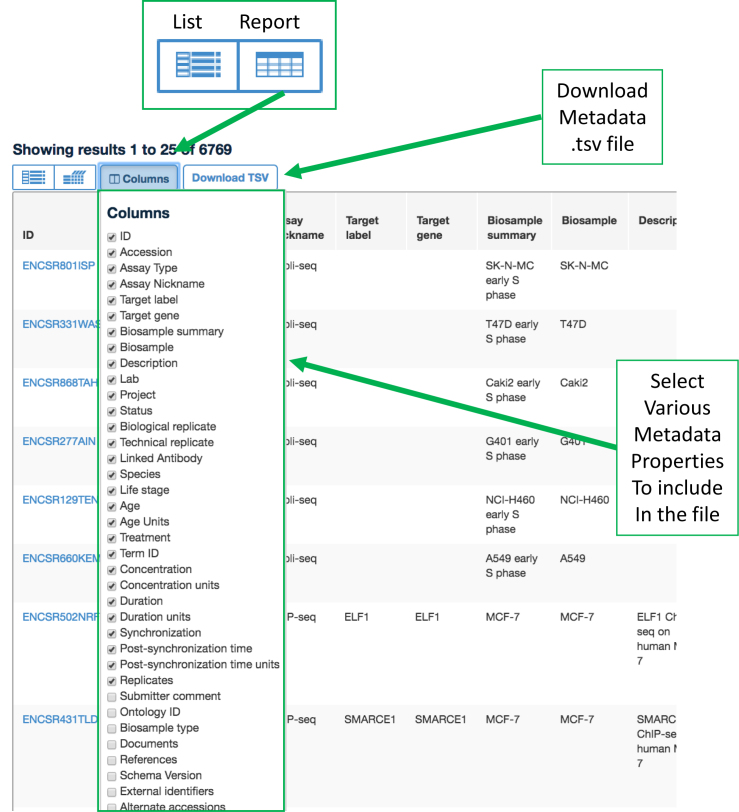
Metadata is captured in JavaScript Object Notation (JSON), a lightweight data interchange format that is easy for humans to read and machines to parse. Metadata tables for datasets can be downloaded in (.tsv) format via the interface (Figure 3) and also obtained programmatically through the REST API (https://www.encodeproject.org/help/rest-api/). These tables are an excellent way to obtain and report the unique accession numbers and accompanying metadata for datasets used in downstream analyses and publications.
Figure 3.
Metadata Download. Illustrating the utility of the ‘reports’ tab to select various metadata properties for data subsets and download that into a table.
New data visualization
The DCC has previously developed methods to create on-the-fly track hubs that can be displayed at the UCSC Genome Browser (10). We have now expanded that feature to the ENSEMBL browser, allowing ready visualization of the data (Figure 4, (11)). Additionally, the DCC has incorporated the BioDalliance Browser (12) which runs as an embedded JavaScript object enabling us to include it in our Portal without the need to link to or rely on external sources.
Figure 4.
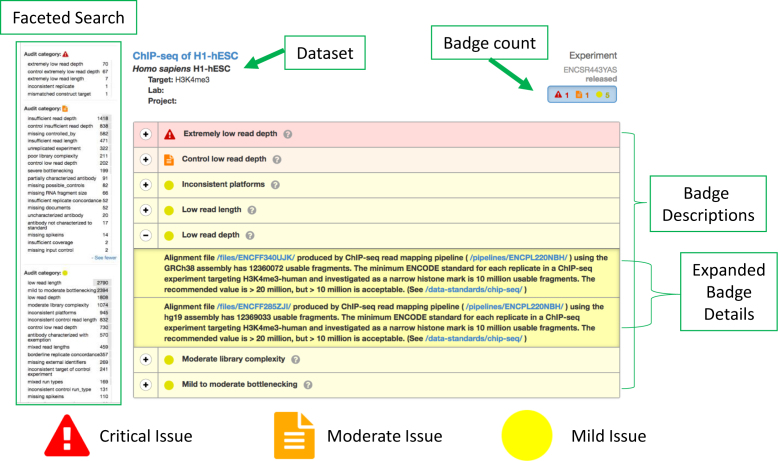
Audits and Badges. Ways in which the automated audits and subsequent badges that are applied to the datasets. This example, shows an experiment as you would see it in the Portal, with its list of color-coded badges. Clicking on the (+) sign, displays a drop-down menu with additional details surrounding each badge and, where available links to the standards and pipelines from which they were determined. Red = a critical issue was identified in the data, orange = a moderate issue was identified in the data, yellow = a mild issue was identified in the data. Additionally, the badge counts are shown under each experiment when displayed in the list-view. The badges have also been incorporated into the faceted search feature on the left side—to enable users to interact with the data in a quality assessed fashion.
Data maintenance, updates and versioning
The data comes from actively funded projects at the DCC and is largely not static - but ‘living’. Data and metadata from the submitting labs is vetted, processed, and released to the public (in most cases) prior to publication (13). The DCC is equipped to rapidly batch reprocess several data types against newer reference genome assemblies as they are made available in addition to accepting data reprocessed externally. We also work with groups to incorporate improved data generated with newer or refined protocols on previously assayed samples. Moreover, as community vetting and ongoing curation of the data take place and issues are identified, the DCC receives requests to change the status (delete, revoke, archive, replaced) of some datasets. Accordingly, we have developed a system to append a ‘status’ to various datasets (Supplementary Table S3) that appears alongside the data. ‘Submitted’ datasets are only visible to the submitting lab and generally in a partial state of compliance pending additional information. ‘Released’ data are available to the public. ‘Revoked’ is used to indicate data that was previously released but over time has proven to be problematic and the submitting labs wish to revoke it from the public sphere. ‘Archived’ is generally used to indicate data that is not problematic but which are not necessarily the most current or up-to-date. ‘Replaced’ is used to consolidate unintentional data duplications (methods described in Gabdank et al., in preparation).
Portal software maintenance, updates and versioning
The genomics field is rapidly moving and consequently the ENCODE Portal is always in an active state of management and development. New data types, pipelines, metadata properties and ways of interacting with the data are continually being devised and improved upon. The Portal has seen many revisions since its launch in 2013 with new software releases occurring on average every three weeks (14). The release version can be found in the lower left corner of the Portal. Labs can keep abreast of the updates by subscribing to or following the DCC communication resources listed in Supplementary Table S4. The software is fully open source and has been adopted by the 4D-Nucleome Project (4DN, (15)) and Clinical Genome Resource (ClinGen, (16)) projects. It is available to download, install, report issues, or pull request at https://github.com/ENCODE-DCC/.
ENCODE PIPELINES
A central focus of the ENCODE project over the last few years has been on building pipelines through which data can be processed in a standardized and centralized fashion. The individual pipelines themselves are built within and arise from the ENCODE production labs. They are then vetted through collaborative working groups and delivered to the DCC. DCC performs extensive testing, hardening, and validation of the software, converting it into a production application. The pipelines are maintained in the DCC GitHub repository (https://github.com/ENCODE-DCC) and have been implemented implements on the DNAnexus (dnanexus.com) cloud platform. The pipelines are therefore accessible to the larger community for unrestricted use on compatible datasets. The DCC has targeted pipelines that support heavily employed assays for currently funded projects, including: RNA-Seq, ChIP-Seq, DNA Accessibility (DNase-Seq and ATAC-Seq assays). The flowcharts outlining the various pipelines the DCC currently has are shown in Supplementary Figures S1–14. A list of URLs on the Portal describing the pipelines can be found in Supplementary Table S2.
Reprocessing of ENCODE data
One advantage of having centralized processing pipelines is that the data can be batch re-processed in a consistent fashion. This avoids the common problem of variation in processing that often add undefined variability in the results. Accordingly, the ENCODE human data that were originally collected and mapped in a non-standard fashion to earlier assemblies in prior project phases has now been reprocessed using the ENCODE unified processing pipelines onto a consistent hg19 (GRCh37) reference genome assembly and the updated GRCh38 reference. Similarly, mouse data previously mapped to mm9 have also been reprocessed onto the latest assembly, mm10. To find data on the Portal mapped against various assemblies navigate to an Experiment page and select the Files tab toggle down, Figure 5). A similar effort is underway for Roadmap Epigenomics data.
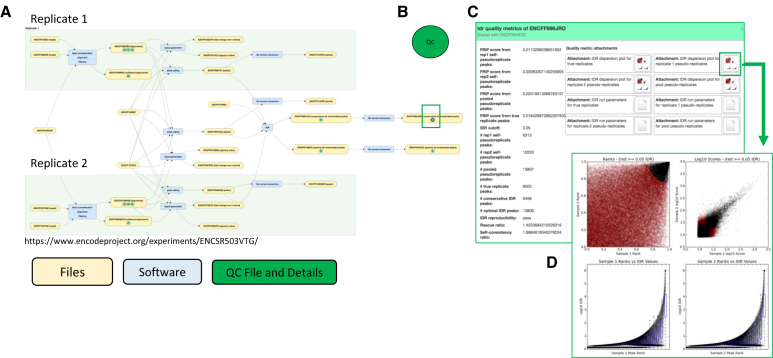
Figure 5.
Pipelines, Provenance and Quality Checks. (A). The File Association Graph for experiment (ENCSR503VTG) is shown. Files and processes specific to each replicate are shown in the green shaded area. Files and processes that are either independent of (genome assembly) or derived form a union of the replicates are shown in the white background. Yellow ovals correspond to accessioned files at the DCC and their accession numbers are all shown. When quality control is run on a given file, a green circle (B). appears and takes users to the quality metrics (C and D). Blue ovals indicate a process done to transform (map, sort, score, etc.) the individual files.
Pipeline quality control metrics
A standard output of all the pipelines includes a basic assessment of quality metrics. These will vary with respect to the particular assay and pipeline but include things such as: mapping rate, percentage of the reads that are unique mappers, Pearson and Spearman correlations of expression values of annotated genes, irreproducible discovery rate (IDR) of peak calls, etc. These values are imported into the Portal and accessible from the File Association Graphs and stored as metadata (Figure 5) and displayed on relevant file pages. Additionally, several of these values are used to determine the automated quality audits that enable us to create the badges that are applied to the datasets.
Modeling of mapping and analysis pipelines
The DCC deals with a variety of data types, some of which we are unable to process through the ENCODE uniform pipelines. In these cases, the production labs will submit both raw and processed data. To ensure that end-users are able to access the computational methods used in the generation of that data—the DCC works with the submitting labs to model the pipeline used to produce the processed files, collecting extensive information on the utilized software tools, versions and parameters underlying each file. These information are initially captured in unstructured PDF files provided by the submitting labs and are appended to the experiments where they are available for download. The information is further culled out and put into structured metadata that is subsequently used to create a File Association Graph (Figure 5) showing the files and the methods through which they are derived, transformed and modified for each experiment.
Mapping reference files
The Portal provides a mechanism to download and track Genome Reference files (and other annotation sets, i.e. gene annotations, spike-in sequences, blacklist regions…) that are bundled into mapping references used by the pipelines. Additionally, the DCC also works with various groups (Genome Reference Consortium (https://www.ncbi.nlm.nih.gov/grc), GA4GH (http://genomicsandhealth.org/), GENCODE (17), etc.) to keep abreast of any changes to reference files be it: versions, file formats, etc. so that we can quickly adapt and modify pipelines accordingly. As with all the data files at the DCC, each distinct reference file gets a unique accession ID that is reported in the File Association Graphs provided on each Experiment page and captured as metadata in the file object JSON, so end-users know which genetic background a given dataset has been processed against (https://www.encodeproject.org/data-standards/reference-sequences/).
Pipeline future directions
As new assays are continually developed and refined, we work with the production labs to co-develop scalable, consistent and robust pipelines that can be implemented in a platform-independent fashion and run on the bolus of their data. Presently, we are working on pipelines and concomitant quality assessments for the following assays: ATAC-Seq, 3D chromosome conformation (Hi-C and ChIA-PET) and to continue to expand upon the existing RNA-Seq, ChIP-Seq and DNase-Seq pipelines to capture additional properties and features applicable to current methods. Efforts are currently underway to develop a generic pipeline framework using Docker and WDL to allow seamless running of ENCODE and other compatible pipelines on any compute cloud or compatible HPC cluster.
ENCODE DATA STANDARDS AND QUALITY CONTROL
To ensure that the data available on the Portal is of high integrity and maximally useful, we have worked with ENCODE Consortium member labs to develop: (i) Experimental guidelines, (ii) Data standards and (iii) Automate data audits and badges to report the overall adherence to the quality standards for each dataset.
Experiment guidelines
The ENCODE Consortium has set up some general experimental guidelines to help unify and streamline data collection and processing within the DCC framework in a high-throughput way. These include things like having two replicates for ChIP-Seq experiments so that peak calling and IDR can be calculated using the DCC pipelines, etc.
Standards
The ENCODE consortium has devised a series of standards for the core data types. The aim of the standards is to provide bins for the automated auditing and badging of the datasets according to certain thresholds. These include values such as: minimum number of reads, minimum read length, ranges of (ir)reproducible scores (IDR, Pearson, Spearman), etc. A current list of data standards can be found at (https://www.encodeproject.org/data-standards/).
Automated audits and badges
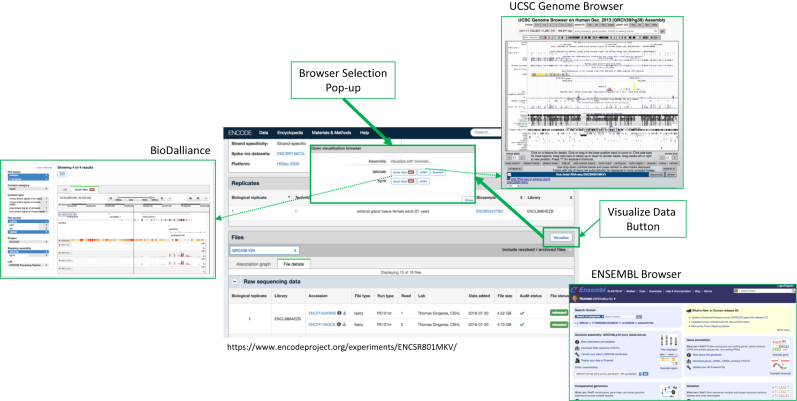
To guide end-users in browsing and selection of datasets, we have developed a system of automated audits that rip through the data and badge it according to: (i) the completion and accuracy of the reported metadata, and (ii) the results of the quality metrics output from the processing pipelines. A current list of audits and badges applied to the datasets audits page (https://www.encodeproject.org/data-standards/audits/). These appear in the Portal alongside each of the datasets (Figure 6), and users can filter the data using the audits tabs in the faceted search function to identify subsets of interest.
Figure 6.
Genome Browsing. ‘Visualize’ link from the experiment pages that takes you to a pop-up menu from which the user can select their preferred Browser (UCSC, ENSEMBL and BioDalliance) and assembly to visualize the data in.
Antibody characterizations
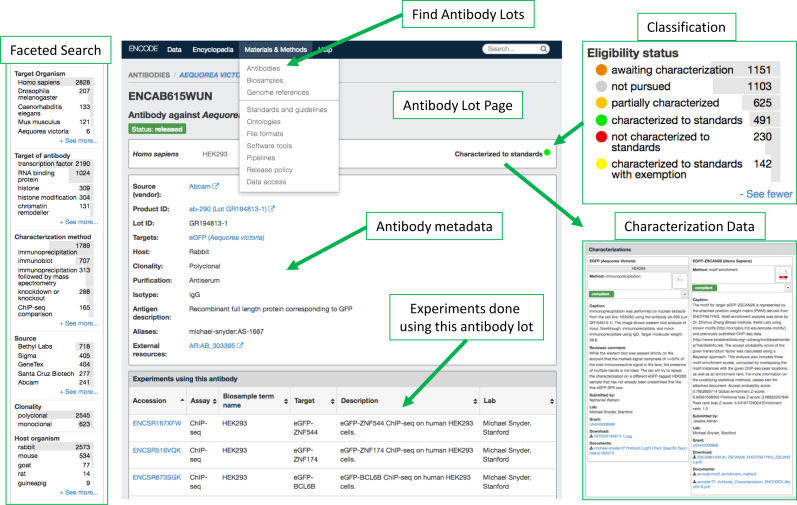
A considerable amount of data on the Portal comes from ChIP-Seq and RNA-binding protein (eCLIP) experiments, assays that rely on antibodies; reagents that can suffer from cross-reactivity, variability in behavior and lack of specificity (18,19). To be able to better assess the specificity of the binding peaks being called, the ENCODE Consortium devoted a large effort towards conducting biochemical antibody characterizations and classifying the data across different metrics depending—in part, on the quality of the antibody characterizations. To date, over 2000 different antibody lots used in the DNA/RNA binding protein and histone modification data collections have been biochemically characterized by the consortium, with the available supporting characterizations for each antibody lot, its review outcome, and its reliant experiments available through the Portal (Figure 7). Together these characterization data are available through the Portal and to the best of our knowledge provide the first ever systematic large-scale assessment of commercial antibodies.
Figure 7.
Antibody characterizations. Example of an Antibody Characterization page for antibody lot (ENCAB615WUN). Antibody lot pages are findable via the drop-down menu. For each lot, various aspects of metadata are displayed including: the Lot number, product, ID, host, targets, etc. At the bottom of the page, the user will find links to experiments done using that lot. Different antibody lots can be found by navigating the faceted search bar on the left. The classification for different lots and its eligibility status for data collection is displayed using a colored dot, the legend of which is shown. Molecular and Biochemical evidence gathered by the labs to test individual lots in various sample types is displayed as well.
FUTURE DIRECTIONS
The DCC continues to be focused on working with groups at the forefront of genomics and technology to develop methods to capture and convey key metadata properties underlying the creation of the early-access data house in the Portal. Future efforts will continue to be focused on identification and analysis of functional elements: (i) increasing and simplifying the user experience; (ii) Working to automate methods to facilitate data deposition; (iii) Pipeline automation and improvements, support for the integrated Encyclopedia of DNA Elements; and (iv) incorporating additional levels of curation and the results of functional testing and screens for active loci under various conditions.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank past and current members of the ENCODE, modENCODE, modERN, GGR and REMC consortia in addition to the staff at the UCSC Genome and ENSEMBL Browsers and the many talented graduate researchers, postdocs, bioinformaticians, etc. for countless hours of data creation, site testing and feedback, etc. for whom this site would not be possible.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Human Genome Research Institute at the National Institutes of Health [U24 HG009397, U41 HG006992]. Funding for open access charge: National Human Genome Research Institute at the United States National Institutes of Health [U24 HG009397].
Conflict of interest statement. None declared.
REFERENCES
- 1. Consortium E.P., Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T. et al. . Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007; 447:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T. et al. . ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016; 44:D726–D732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosenbloom K.R., Dreszer T.R., Pheasant M., Barber G.P., Meyer L.R., Pohl A., Raney B.J., Wang T., Hinrichs A.S., Zweig A.S. et al. . ENCODE whole-genome data in the UCSC genome browser. Nucleic Acids Res. 2010; 38:D620–D625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roadmap Epigenomics Consortium Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al. . Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bujold D., Morais D.A., Gauthier C., Cote C., Caron M., Kwan T., Chen K.C., Laperle J., Markovits A.N., Pastinen T. et al. . The International Human Epigenome Consortium Data Portal. Cell Syst. 2016; 3:496–499. [DOI] [PubMed] [Google Scholar]
- 7. Mod E.C., Roy S., Ernst J., Kharchenko P.V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L. et al. . Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010; 330:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mouse E.C., Stamatoyannopoulos J.A., Snyder M., Hardison R., Ren B., Gingeras T., Gilbert D.M., Groudine M., Bender M., Kaul R. et al. . An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 2012; 13:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerstein M.B., Lu Z.J., Van Nostrand E.L., Cheng C., Arshinoff B.I., Liu T., Yip K.Y., Robilotto R., Rechtsteiner A., Ikegami K. et al. . Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010; 330:1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T., Nguyen N., Paten B., Zweig A.S., Karolchik D. et al. . Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014; 30:1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al. . Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Down T.A., Piipari M., Hubbard T.J.. Dalliance: interactive genome viewing on the web. Bioinformatics. 2011; 27:889–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hong E.L., Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M. et al. . Principles of metadata organization at the ENCODE data coordination center. Database (Oxford). 2016; 2016:baw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hitz B.C., Rowe L.D., Podduturi N.R., Glick D.I., Baymuradov U.K., Malladi V.S., Chan E.T., Davidson J.M., Gabdank I., Narayana A.K. et al. . SnoVault and encodeD: a novel object-based storage system and applications to ENCODE metadata. PLoS One. 2017; 12:e0175310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dekker J., Belmont A.S., Guttman M., Leshyk V.O., Lis J.T., Lomvardas S., Mirny L.A., O’Shea C.C., Park P.J., Ren B. et al. . The 4D nucleome project. Nature. 2017; 549:219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L. et al. . ClinGen–the Clinical Genome Resource. N. Engl. J. Med. 2015; 372:2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. . GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baker M. Reproducibility crisis: blame it on the antibodies. Nature. 2015; 521:274–276. [DOI] [PubMed] [Google Scholar]
- 19. Weller M.G. Quality issues of research antibodies. Anal. Chem. Insights. 2016; 11:21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.