Abstract
For more than 30 years, the International Nucleotide Sequence Database Collaboration (INSDC; http://www.insdc.org/) has been committed to capturing, preserving and providing access to comprehensive public domain nucleotide sequence and associated metadata which enables discovery in biomedicine, biodiversity and biological sciences. Since 1987, the DNA Data Bank of Japan (DDBJ) at the National Institute for Genetics in Mishima, Japan; the European Nucleotide Archive (ENA) at the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) in Hinxton, UK; and GenBank at National Center for Biotechnology Information (NCBI), National Library of Medicine, National Institutes of Health in Bethesda, Maryland, USA have worked collaboratively to enable access to nucleotide sequence data in standardized formats for the worldwide scientific community. In this article, we reiterate the principles of the INSDC collaboration and briefly summarize the trends of the archival content.
INTRODUCTION
The International Nucleotide Sequence Database Collaboration (1) (INSDC: http://www.insdc.org) represents one of the most celebrated global initiatives in public domain data sharing. The collaboration consists of three nodes: DNA Data Bank of Japan (DDBJ: http://www.ddbj.nig.ac.jp/) in Mishima, Japan (2); European Nucleotide Archive (ENA: http://www.ebi.ac.uk/ena/) in Hinxton, UK (3) and GenBank (https://www.ncbi.nlm.nih.gov/genbank/) in Bethesda, Maryland, USA (4). The INSDC members work together to ensure that all public domain nucleotide sequence data deposited in the archives is preserved as part of the scientific record and is accessible in standardized formats across the three sites through daily data exchange. The INSDC archives work together to respond to emerging sequencing technologies. The scope of data in INSDC includes raw sequence reads and alignments in the read archives (SRA), and assembled sequences with functional annotation in the traditional archives. Structured metadata describing the biological sample including taxonomic information, experimental design and project scope are submitted along with the sequences to provide context. The INSDC works in concert with appropriate standards communities, such as the Genomics Standards Consortium for environmental microbiology data (5) and the Global Microbial Identifier for pathogen data (http://www.globalmicrobialidentifier.org/) to ensure rich metadata capture for understanding the origin of the sequences. Each center provides tools to facilitate the deposition of data and associated metadata, as well as gateways for the analysis and retrieval of deposited data. Routine data exchange through standardized formats provides global synchrony across the collaboration to facilitate the study of living things through sequence analysis. These long-held tenets of INSDC are a model for the FAIR Data Principles (6) which promotes published data to be Findable, Accessible, Interoperable and Reusable.
COLLABORATION
Members of the INSDC meet annually to discuss issues related to building and maintaining the sequence archives. The database standards and policies that result from these meetings are presented on the INSDC website (http://www.insdc.org/). The Feature Table Definitions Document (http://www.insdc.org/documents/feature-table) describes feature keys and qualifiers presented in the Flat File report format in the traditional archives. Many of the feature key qualifiers use controlled vocabularies (http://www.insdc.org/insdc-controlled-vocabularies). In addition, documents are provided that describe policies for acceptance of certain data types such as genome assemblies and Third Party (TPA), and best practices for data deposited to the public archives (http://www.insdc.org/documents).
Each center provides its user community with tools for the submission of nucleotide sequence data. Improvements are being made to submissions systems at all three sites to make submitting data easier through templated web wizards that guide the submitter to provide rich contextual information along with the sequences and annotation. Validations within the wizards ensure that minimal requirements have been met and that the data are syntactically and semantically valid. A submitter deposits their data at one site and through a coordinated exchange, the data will be presented at all three sites.
Each center also provides its user community with tools for the retrieval and analysis of the sequenced data. Though each center has its own tools, the data presented at each site is the same due to the nightly exchange of data. Sequences are accessioned across a single namespace such that an accession search yields the same data content regardless of where the data are accessed.
POLICY
INSDC data are provided openly and free of charge to users. Data presented in the archive can be retrieved and incorporated in subsequent studies which may lead to important scientific discoveries. Ciiting INSDC accession numbers associated with each sequence ensures that the original data submitter is properly credited in accordance with FAIR data sharing principles.
Submitters to the database may request that their sequence records are made publicly available immediately following submission. Alternatively, a submission may be kept confidential prior to publication but data are released publicly as soon as the work is presented in a publication.
To comply with the consent agreements of human donors who have provided material for sequencing, authorization may be required for access to this data. INSDC archives do not manage these records, rather each partner's institute works under their respective legislative systems with the appropriate ethical bodies and committees to implement appropriate levels of security in their respective data archives (JGA at DDBJ (2); EGA at EMBL-EBI (7) and dbGaP at NCBI (8).
INSDC databases are data hosts and not owners; data ownership, and hence editorial control of the scientific content, remains with the original data provider. However, during submission processing, database staff may make minor modifications to submitted data in an effort to provide standardized, validated records to the users. Furthermore, only data owners and their approved delegates are permitted to update their records. To ensure consistency, updates to data must be performed at the INSDC node where the data was initially submitted. The updated records are then propagated to the partner nodes.
As a requirement for publication in most journals, any new sequence described in the article should be submitted to INSDC and the accession numbers assigned to the data be cited in the article. In the past, the INSDC worked with journal editors to establish this policy so that that a reader will have access to the underlying data that was described in the paper. In 2016, this principle of data sharing and data citation was reaffirmed by the International Advisory Committee for INSDC in a letter to the scientific community (9,10).
CONTENT IN 2017
Since our previous report on the status of the International Nucleotide Sequence Database Collaboration (1), the assembled/annotated portion of the sequence data maintained by the INSDC grew from 1.432 trillion bases in August 2015 to 2.650 trillion bases in August 2017. The growth rate over this two-year period was 185%, just short of a doubling. During the same time period, the read archive grew by 233% with the addition of 3000 trillion bases. The space required to store a single copy of the reads, increased from 1.5 to 3.2 Petabytes, an increase of 210%. Storage efficiency increased due to storing a greater fraction of submitted data in aligned and compressed format.
The cumulative growth in the number of sequenced bases and the number of sequence records in the assembled/annotated portion of the archive over the last decade is detailed in Figure 1A. The doubling time for the number of bases during this ten-year period is 28.4 months, while the doubling time for the number of records is 37.9 months. Figure 1B depicts the growth of the read archive both in storage and in space.
Figure 1.
(A) Cumulative 10-Year INSDC Growth of Assembled/Annotated Data: Sequence bases (solid) and sequence records (dashed). (B) Cumulative 10-Year INSDC Growth of SRA Data: Sequence bases (solid) and single-copy data storage (dashed).
TAXONOMY
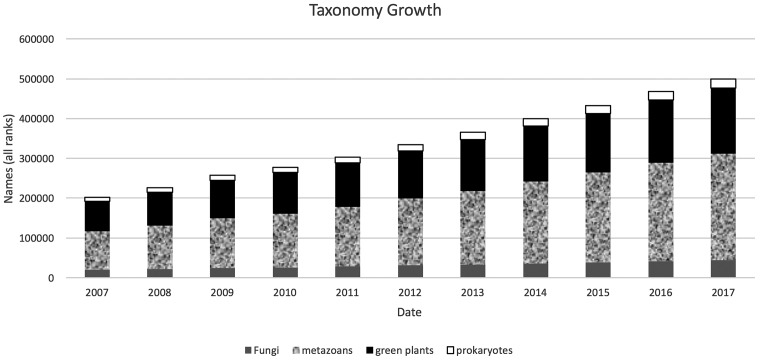
The NCBI Taxonomy database (http://www.ncbi.nlm.nih.gov/taxonomy) provides a central organizing hub for many INSDC resources and is curated by taxonomist specialists residing at NCBI (11). All partners in the INSDC send consults whenever a sequence is submitted with an organism name that is not present in the taxonomy database. This originated from a 1997 agreement by the INSDC members to resolve taxonomic issues prior to the release of new sequence data. The final number for all taxa on 1 January 2017 was 512 941. The yearly increase of formal taxonomic names used by INSDC is indicated in Figure 2. The viruses and non-metazoan eukaryotes (4194 and 8940 names respectively on 1 January 2017) are not indicated. It is evident that the increase is constant in all cases with the ratio between the different groups very stable as well.
Figure 2.
Cumulative 10-Year INSDC Growth of Formal Taxonomic Names (all ranks). Names are broken down into Fungi, metazoan eukaryotes (Metazoa), green plants (Viridiplantae) and prokaryotes (Archaea combined with Bacteria). For simplicity, non-metazoan eukaryotic groups and viruses are excluded.
In addition to cataloguing taxonomic names, the database also keeps track of voucher identifiers assigned by museums, herbariums and other collections that are declared as type material. This is usually a physical specimen or culture that was assigned during the formal process of describing new species names under rules defined by the various codes of biological nomenclature (12). These vouchers have a special status and are crucial for any comparative work done to determine species identity. Genome sequences derived from type material are currently used to correct the taxonomic assignments of prokaryotic genomes at NCBI. A new INSDC qualifier (/type_material) was introduced and will be added to INSDC records using information from the NCBI Taxonomy database. A list of accepted terms that describes the classes of type material is available from http://www.insdc.org/controlled-vocabulary-typematerial-qualifer
FUTURE OUTLOOK
While a variety of high-throughput life science assay platforms are emerging into the ‘big data’ limelight, we expect that interest in nucleic acid sequencing will continue to grow and be adopted by broader user communities. With ever higher yields and increasing affordability, nucleic acid sequencing is adopted for new uses and to supplement other biological assay types. We see growth in community and population genomics, metagenomes and whole biome microbial surveys like the TARA oceans project (13). Such large-scale efforts not only expand the breadth and depth of scientific knowledge but yield actionable insights valuable to medicine, crop and livestock industries. Sequencing is also proving its value to clinical diagnostics and microbial pathogen surveillance (14). We expect nucleic acid sequence submissions and the need for re-analysis and re-use to continue to grow across existing and new user communities.
Given the expected onward growth, INSDC partners continue their development and maintenance of scalable data submission and retrieval systems. The INSDC assures uniform and synchronized data content. Technical implementation details and software development are managed by each partner in accordance with their stakeholders and host institutions.
We will continue to engage with international initiatives and the life-sciences community as they drive application of sequencing in different domains. With the increasing value of sequence data and the time-sensitive nature of pathogen surveillance, we continue our work with the Global Microbial Identifier (GMI) initiative in building a global system for rapid sharing of well-structured whole genome sequence data across bacteria, viruses and eukaryotic parasites.
The INSDC is committed to providing well-described data sets with maximum discoverability, interoperability and reusability. To this end, we work extensively with community standards groups like the Genomics Standards Consortium on integrating the MIxS standards into submission procedures which yields rich, yet practical, checklist or package based metadata standards for organismal and metagenomic data sets (15). In addition, we lead the Data Standards Working Group of the GMI that drives at metadata and data standards around shared whole genome pathogen sequencing data.
Finally, while we will continue to enjoy financial support from our respective institutions, organizations and regional funders, we are actively participating in the global effort to develop a sound footing for core bioinformatics resources, through the HSFPO initiative (16).
FUNDING
NCBI by the Intramural Research Program of the National Institutes of Health; National Library of Medicine; European Nucleotide Archive by the European Molecular Biology Laboratory, the Horizon 2020 Programme of the European Commission and the Biotechnology and Biological Sciences Research Council; DDBJ by the Ministry of Education, Culture, Sports, Science and Technology, the Research Organization of Information and Systems, Japan. Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cochrane G., Karsch-Mizrachi I., Takagi T.. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2016; 44:D48–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mashima J., Kodama Y., Fujisawa T., Katayama T., Okuda Y., Kaminuma E., Ogasawara O., Okubo K., Nakamura Y., Takagi T.. DNA Data Bank of Japan. Nucleic Acids Res. 2017; 45:D25–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toribio A.L., Alako B., Amid C., Cerdeno-Tarraga A., Clarke L., Cleland I., Fairley S., Gibson R., Goodgame N., Ten Hoopen P. et al. . European Nucleotide Archive in 2016. Nucleic Acids Res. 2017; 45:D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2017; 45:D37–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Field D., Sterk P., Kottmann R., De Smet J.W., Amaral-Zettler L., Cochrane G., Cole J.R., Davies N., Dawyndt P., Garrity G.M. et al. . Genomic standards consortium projects. Standards Genomic Sci. 2014; 9:599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E. et al. . The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lappalainen I., Almeida-King J., Kumanduri V., Senf A., Spalding J.D., Ur-Rehman S., Saunders G., Kandasamy J., Caccamo M., Leinonen R. et al. . The European Genome-phenome Archive of human data consented for biomedical research. Nat. Genet. 2015; 47:692–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tryka K.A., Hao L., Sturcke A., Jin Y., Wang Z.Y., Ziyabari L., Lee M., Popova N., Sharopova N., Kimura M. et al. . NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014; 42:D975–D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blaxter M., Danchin A., Savakis B., Fukami-Kobayashi K., Kurokawa K., Sugano S., Roberts R.J., Salzberg S.L., Wu C.I.. Reminder to deposit DNA sequences. Science. 2016; 352:780. [DOI] [PubMed] [Google Scholar]
- 10. Salzberg S.L. Databases: Reminder to deposit DNA sequences. Nature. 2016; 533:179. [DOI] [PubMed] [Google Scholar]
- 11. Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012; 40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Federhen S. Type material in the NCBI Taxonomy Database. Nucleic Acids Res. 2015; 43:D1086–D1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karsenti E., Acinas S.G., Bork P., Bowler C., De Vargas C., Raes J., Sullivan M., Arendt D., Benzoni F., Claverie J.M. et al. . A holistic approach to marine eco-systems biology. PLoS Biol. 2011; 9:e1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allard M.W., Strain E., Melka D., Bunning K., Musser S.M., Brown E.W., Timme R.. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J. Clin. Microbiol. 2016; 54:1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yilmaz P., Kottmann R., Field D., Knight R., Cole J.R., Amaral-Zettler L., Gilbert J.A., Karsch-Mizrachi I., Johnston A., Cochrane G. et al. . Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 2011; 29:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson W.P. Data management: A global coalition to sustain core data. Nature. 2017; 543:179. [DOI] [PubMed] [Google Scholar]