Abstract
Background and objectives
Steroid-resistant nephrotic syndrome overwhelmingly progresses to ESRD. More than 30 monogenic genes have been identified to cause steroid-resistant nephrotic syndrome. We previously detected causative mutations using targeted panel sequencing in 30% of patients with steroid-resistant nephrotic syndrome. Panel sequencing has a number of limitations when compared with whole exome sequencing. We employed whole exome sequencing to detect monogenic causes of steroid-resistant nephrotic syndrome in an international cohort of 300 families.
Design, setting, participants, & measurements
Three hundred thirty-five individuals with steroid-resistant nephrotic syndrome from 300 families were recruited from April of 1998 to June of 2016. Age of onset was restricted to <25 years of age. Exome data were evaluated for 33 known monogenic steroid-resistant nephrotic syndrome genes.
Results
In 74 of 300 families (25%), we identified a causative mutation in one of 20 genes known to cause steroid-resistant nephrotic syndrome. In 11 families (3.7%), we detected a mutation in a gene that causes a phenocopy of steroid-resistant nephrotic syndrome. This is consistent with our previously published identification of mutations using a panel approach. We detected a causative mutation in a known steroid-resistant nephrotic syndrome gene in 38% of consanguineous families and in 13% of nonconsanguineous families, and 48% of children with congenital nephrotic syndrome. A total of 68 different mutations were detected in 20 of 33 steroid-resistant nephrotic syndrome genes. Fifteen of these mutations were novel. NPHS1, PLCE1, NPHS2, and SMARCAL1 were the most common genes in which we detected a mutation. In another 28% of families, we detected mutations in one or more candidate genes for steroid-resistant nephrotic syndrome.
Conclusions
Whole exome sequencing is a sensitive approach toward diagnosis of monogenic causes of steroid-resistant nephrotic syndrome. A molecular genetic diagnosis of steroid-resistant nephrotic syndrome may have important consequences for the management of treatment and kidney transplantation in steroid-resistant nephrotic syndrome.
Keywords: genetic renal disease; pediatric; molecular genetics; Child; Humans; Nephrosis, congenital; nephrotic syndrome; Exome; kidney transplantation; Mutation; Kidney Failure, Chronic; Renal Insufficiency, Chronic; Phenotype
Introduction
Nephrotic syndrome in childhood is characterized by proteinuria (>40 mg/m2 per hour), hypoalbuminemia, edema, and hyperlipidemia. It can cause hypertension, severe infections, and thrombotic events. Patients are classified by their response to steroid therapy. In children and young adults, about 80% of patients respond to standard steroid therapy and are termed steroid sensitive (1). In contrast, individuals with steroid-resistant nephrotic syndrome overwhelmingly progress to CKD and ESRD. At this time, there is no effective therapy to curtail the relentless progression to ESRD.
The most frequent kidney histologic feature of steroid-resistant nephrotic syndrome is FSGS. In patients with FSGS, the risk of recurrence after kidney transplantation is estimated to be approximately 33% (2–4). FSGS constitutes the third most prevalent cause of ESRD in the first two decades of life (5). To date, >30 monogenic causes of steroid-resistant nephrotic syndrome have been identified (6), many of which implicate the glomerular podocyte and slit membrane as the primary sites where the pathogenesis of steroid-resistant nephrotic syndrome unfolds (7). The majority of genes known to cause steroid-resistant nephrotic syndrome are recessively inherited. Patients with mutations in these genes manifest with steroid-resistant nephrotic syndrome in childhood and adolescence, whereas dominant steroid-resistant nephrotic syndrome genes often manifest later in life.
We showed previously by targeted panel sequencing of 27 known steroid-resistant nephrotic syndrome genes that in 30% of steroid-resistant nephrotic syndrome cases with onset before 25 years, a causative mutation can be detected (8). However, panel sequencing by multiplex PCR is limited by requiring large numbers of Sanger sequencing to confirm individual genetic variants (8). Additionally, evaluation of genes by panel sequencing is limited to approximately 30 genes. With the growing number of genes available, we sought a mechanism by which we could evaluate a patient for a high number of steroid-resistant nephrotic syndrome genes, as well as detect novel causes of nephrosis should no known gene be identified.
In a cohort of patients with CKD and the phenotype of increased kidney echogenicity, we identified a causative mutation in 63% using whole exome sequencing (9). We evaluated here the utility of whole exome sequencing in an international cohort with steroid-resistant nephrotic syndrome. To date, this cohort is the largest to undergo whole exome sequencing (10). Given the very high rate of establishing an etiologic diagnosis and the significant implications for clinical management and pretransplant and post-transplant care, whole exome sequencing should be considered in all individuals with steroid-resistant nephrotic syndrome diagnosed before age 25 years.
Materials and Methods
Human Participants
The study was approved by the institutional review board of the University of Michigan and Boston Children’s Hospital. From April of 1998 to June of 2016, patients were enrolled after obtaining informed consent. Inclusion criteria were: onset of symptoms before 25 years and a clinical diagnosis of steroid-resistant nephrotic syndrome (e.g., proteinuria, hypoalbuminemia, edema) or nephrotic range proteinuria with kidney histology of FSGS or diffuse mesangial sclerosis (Supplemental Table 1). Three hundred thirty-five individuals from 300 families were enrolled. Before December of 2013, enrolled individuals were screened for mutations in WT1 and NPHS2. Those that screened positive were not included in this study.
Whole Exome Sequencing and Variant Calling
Whole exome sequencing and variant burden analysis were performed as previously described (11–13). Genomic DNA was isolated from blood lymphocyte or saliva samples and subjected to exome capture using Agilent SureSelect human exome capture arrays (Life technologies) followed by next generation sequencing on the Illumina HighSeq sequencing platform. Sequence reads were mapped to the human reference genome assembly (NCBI build 37/hg19) using CLC Genomics Workbench (version 6.5.2) software (CLC bio, Aarhus, Denmark). After alignment to the human reference genome (GRCh37/hg19), variants were filtered for most likely nondeleterious variants as previously described (8,11). Variants with minor allele frequencies >1% in the dbSNP (version 142) or in the 1000 Genomes Project (1094 patients of various ethnicities; May 2011 data release) databases were excluded because they were unlikely to be deleterious. We used manual inspection for the p.Arg229Gln mutation in the NPHS2 gene which is reported to be deleterious with other variants, which would be filtered out using this method (14). Synonymous variants and intronic variants that were not located within splice site regions were excluded. Remaining variants included nonsynonymous variants and splice site variants.
Mutation Calling in Known Steroid-Resistant Nephrotic Syndrome Genes
We evaluated whole exome sequencing data for causative mutations in 33 steroid-resistant nephrotic syndrome genes known at the time (Supplemental Table 2). Mutation calling was applied as stated above, followed by filtering remaining variants for changes in the regions of the 33 genes. Remaining variants were ranked on the basis of their probable effect on the function of the encoded protein considering evolutionary conservation among orthologs using ENSEMBL Genome Browser and assembled using Clustal Omega, as well as PolyPhen-2 (15), SIFT (16), and MutationTaster (17). Mutations were designated as likely pathogenic on the basis of criteria given by Supplemental Table 3. Mutation calling was performed by clinician scientists/geneticists, with knowledge of the clinical phenotypes and pedigree structure, as well as experience with homozygosity mapping and whole exome sequencing evaluation. Remaining variants were confirmed in patient DNA by Sanger sequencing as previously described (8). Whenever parental DNA was available, segregation analysis was performed.
If no causative mutation was identified, we evaluated for mutations in genes that may represent phenocopies of steroid-resistant nephrotic syndrome (Supplemental Table 2). Variants were evaluated as above. Correlation of genotype and phenotype was examined and, if matching, the genetic variant was deemed a causative mutation.
Mutation Calling to Identify Novel Causes of Steroid-Resistant Nephrotic Syndrome
If no causative mutation was found in a known steroid-resistant nephrotic syndrome gene and a family had homozygosity (>100 Mbp) detected after homozygosity mapping, we then evaluated whole exome sequencing data for homozygous variants. Single heterozygous variants were excluded. We applied homozygosity mapping in consanguineous families or linkage analysis in sibling cases to filter variants (18). Remaining variants were ranked as described above. Variants were confirmed as described above.
Homozygosity Mapping and Linkage Analysis
Before 2014, for genome-wide homozygosity mapping, the GeneChip Human Mapping 250k d Array (Affymetrix) was used. Alternatively, homozygosity mapping data were generated from whole exome sequencing data. Nonparametric logarithm (base 10) of odds scores were calculated using a modified version of the program GENEHUNTER2.1 (19,20) through stepwise use of a sliding window with sets of 110 SNPs and the program ALLEGRO (21) in order to identify regions of homozygosity as described (18,22) using a disease allele frequency of 0.0001 and white marker allele frequencies. To generate homozygosity mapping after 2014, downstream processing of aligned binary alignment map files was done using Picard and SAMtools4 (23). Single nucleotide variants calling was performed using Genome Analysis Tool Kit (24) and the generated variant call format file was subsequently used in Homozygosity Mapper (25).
Web Resources
UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway;
1000 Genomes Browser, http://browser.1000genomes.org;
Clustal Omega, http://www.ebi.ac.uk/Tools/msa/clustal;
Ensembl Genome Browser, http://www.ensembl.org;
Exome Variant Server, http://evs.gs.washington.edu/EVS;
Exome Aggregation Consortium, exac.broadinstitute.org;
HGMD Professional 2016.3, https://portal.biobase-international.com/hgmd;
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org;
Polyphen2, http://genetics.bwh.harvard.edu/pph2;
Sorting Intolerant From Tolerant (SIFT), http://sift.jcvi.org;
MutationTaster, http://www.mutationtaster.org.
Results
Identification of Causative Mutations in One of 20 Steroid-Resistant Nephrotic Syndrome Genes in 25% of Steroid-Resistant Nephrotic Syndrome Cases
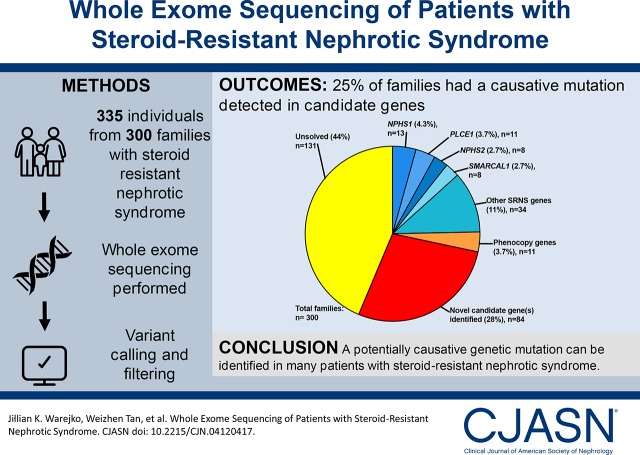
Whole exome sequencing was performed in 335 individuals from 300 families and evaluated for mutations in the 33 steroid-resistant nephrotic syndrome genes known at the time (Table 1). In 74 families (25%), a causative mutation in one of 20 known steroid-resistant nephrotic syndrome genes was detected (Figure 1, Table 1). NPHS1 (13 families), PLCE1 (11 families), NPHS2 (eight families), and SMARCAL1 (eight families) were the most common genes in which mutations were identified, comprising 51% of all mutations identified (Figure 1, Table 1).
Table 1.
Number and proportion of 300 families with steroid-resistant nephrotic syndrome, in whom causative mutations in one of 23 known monogenic causes of steroid-resistant nephrotic syndrome were detected
| Gene | Number of Families with Causative Mutation | Percentage of Families with Gene, % | Number of Mutations Known from Biobasea | Number of Novel Mutations |
|---|---|---|---|---|
| SRNS genes | ||||
| NPHS1 | 13 | 18 | 10 | 1 |
| PLCE1 | 11 | 15 | 8 | 2 |
| NPHS2 | 8 | 11 | 9 | 1 |
| SMARCAL1 | 8 | 11 | 5 | 1 |
| LAMB2 | 6 | 8 | 3 | 3 |
| NUP93 | 4 | 5 | 2 | 1 |
| MYO1E | 2 | 3 | 1 | 1 |
| SGPL1 | 3 | 4 | — | 2 |
| WDR73 | 3 | 4 | 3 | — |
| ITGA3 | 2 | 3 | 2 | — |
| LMX1B | 2 | 3 | 1 | — |
| NUP205 | 2 | 3 | 2 | — |
| TTC21B | 2 | 3 | 1 | 1 |
| WT1 | 2 | 3 | 1 | — |
| COQ2 | 1 | 1 | 1 | 1 |
| DGKE | 1 | 1 | 1 | — |
| INF2 | 1 | 1 | — | 1 |
| KANK4 | 1 | 1 | 1 | — |
| PDSS2 | 1 | 1 | 1 | — |
| TRPC6 | 1 | 1 | 1 | — |
| Total | 74 | 100 | 53 | 15 |
| Phenocopy genes | ||||
| COL4A5 | 3 | 27 | 2 | — |
| AGXT | 2 | 18 | 0 | 2 |
| CLCN5 | 1 | 9.1 | — | 1 |
| COL4A3 | 1 | 9.1 | — | 1 |
| CTNS | 1 | 9.1 | 1 | — |
| FN1 | 1 | 9.1 | — | 1 |
| GLA | 1 | 9.1 | 1 | — |
| WDR19 | 1 | 9.1 | 1 | — |
| Total | 11 | 100.0 | 5 | 5 |
Fifty-three of the mutations detected have previously been reported in BioBase, and 15 are novel (respective genes are under the table subheading ‘SRNS genes’). The most common genes to have a mutation detected in steroid-resistant nephrotic syndrome families were NPHS1, PLCE1, NPHS2, and SMARCAL1 (51% of all steroid-resistant nephrotic syndrome gene mutations detected). In an additional 11 families, mutations were detected in eight genes that cause a kidney disease that is a phenocopy of steroid-resistant nephrotic syndrome (respective genes are under the table subheading ‘Phenocopy genes’). Five of the mutations identified have previously been reported in BioBase, and five are novel. SRNS, steroid-resistant nephrotic syndrome; —, no mutations detected.
Figure 1.
In 74 of 300 (25%) families with steroid-resistant nephrotic syndrome, a causative mutation was detected in one of 20 genes known to cause steroid-resistant nephrotic syndrome (shades of blue). In 3.7% of families, a mutation was found in genes causing a kidney disease that may represent phenocopies of steroid-resistant nephrotic syndrome (orange). In 28% of families, one or more potential novel candidate genes were identified (red). In 44% of families, no causative mutations or candidate genes were detected. SRNS, steroid-resistant nephrotic syndrome.
Ninety-four of the 300 families studied by whole exome sequencing have been previously studied using Fluidigm panel sequencing. The overlap between cohorts is given in Supplemental Table 4. We found that, whereas in 20 of 74 (27%) families the causative mutation was previously detected using panel sequencing, 9 of 74 (12%) had a diagnosis made by whole exome sequencing and not by panel sequencing. In an additional 28% of families, we detected a likely causative mutation in one or more potential novel steroid-resistant nephrotic syndrome genes (Figure 1), given in Supplemental Table 5.
Novel Mutations Detected in Known Steroid-Resistant Nephrotic Syndrome Genes
We detected 68 different mutations in 20 of 33 known steroid-resistant nephrotic syndrome genes, 53 of which had previously been reported in the literature (Table 1). Fifteen novel mutations have not been reported previously (Table 1). Individual families in whom a causative mutation was detected are described in Supplemental Table 9.
Whole Exome Sequencing Identifies Phenocopies in 11 of 90 Families with a Causative Mutation Detected
We detected a causative mutation in 11 of 300 families with steroid-resistant nephrotic syndrome (3.7%) (Figure 1, Table 1). Mutations were found in eight phenocopy genes, specifically COL4A5, COL4A3, CLCN5, GLA, AGXT, CTNS, FN1, and WDR19. A total of ten different mutations were detected, five of which are novel (Table 1).
Novel Candidate Genes Are Identified Using Whole Exome Sequencing
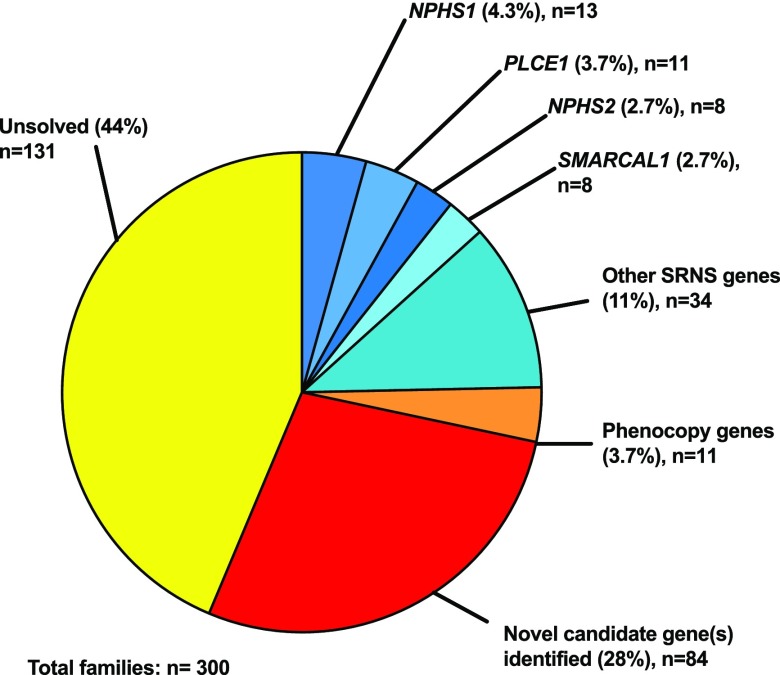
In 61 of 146 (42%) consanguineous families with no causative mutation found in a known steroid-resistant nephrotic syndrome gene, one or more candidate genes were detected using homozygosity mapping (Figure 2, Supplemental Table 5). In nonconsanguineous families with >1 individual affected, linkage analysis was used to identify a potentially causative mutation in 18 of 135 families (13%).
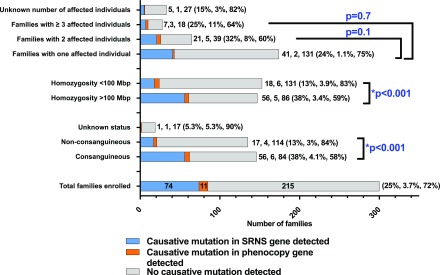
Figure 2.
A causative mutation in a steroid-resistant nephrotic syndrome gene, phenocopy gene, or novel candidate gene is detected in a greater proportion of consanguineous families compared to non-consanguineous families. We detected a causative mutation in 38% of consanguineous families and 13% of nonconsanguineous families. Through homozygosity mapping and a recessive hypothesis, we were able to identify potentially causative mutations in 42% of consanguineous families. Potential causative mutations in novel candidate genes were detected in nonconsanguineous families by evaluating for overlapping genes in siblings. Percentages>10% are rounded to the nearest whole number. SRNS, steroid-resistant nephrotic syndrome.
Description of Cohort
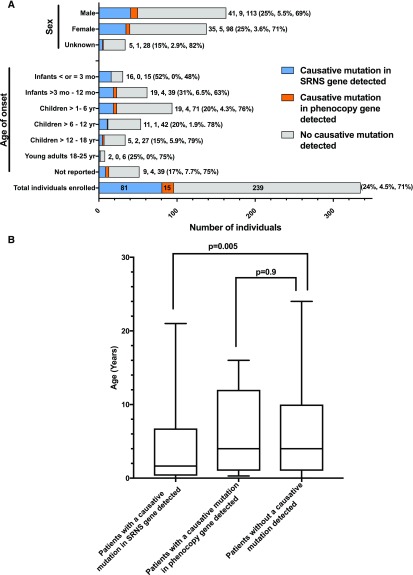
Onset of illness ranged from birth to 24 years of age (Figure 3A, Supplemental Table 6). The median age in individuals in whom a causative mutation was detected in a steroid-resistant nephrotic syndrome gene was 1.7 years versus 4 years in those without a causative mutation identified, which was statistically significant (Figure 3B).
Figure 3.
After dividing the 335 individuals from 300 families with steroid-resistant nephrotic syndrome by gene identification status (steroid-resistant nephrotic syndrome gene, phenocopy gene, no mutation detected) and sorting by age and sex, the median age of individuals with a mutation detected in a steroid resistant nephrotic syndrome gene is significantly lower than the median age of individuals with a mutation detected in a phenocopy gene or individuals with no mutation detected. (A) Families in whom a causative mutation in a known steroid-resistant nephrotic syndrome gene (blue) or phenocopy gene (orange) was detected as compared with those families where no causative mutation was detected (gray). Bars and numbers at end of bars represent number of affected individuals in each category, divided into those with a causative mutation detected in a steroid-resistant nephrotic syndrome gene (blue), those with a causative mutation detected in a phenocopy gene (orange), and those without a causative mutation detected (gray). Percentages at end of each bar reflect the same three categories. Percentages>10% are rounded to the nearest whole number. (B) Median age of onset in patients with a causative mutation detected in a steroid-resistant nephrotic syndrome gene was 1.7 years versus 4 years in those without a mutation detected (range, 0–24 years). For those with a causative mutation detected in a steroid-resistant nephrotic syndrome gene, the range was 0–21 years. Mann–Whitney U test P<0.01. Median age of individuals with a phenocopy mutation detected was 4 years (range, 0.3–16), which was not statistically significant. Data of the characteristics of the steroid-resistant nephrotic syndrome cohort compared with the subcohort of those individuals with a causative mutation detected in a steroid-resistant nephrotic syndrome gene or phenocopy gene are given in Supplemental Table 3. SRNS, steroid-resistant nephrotic syndrome.
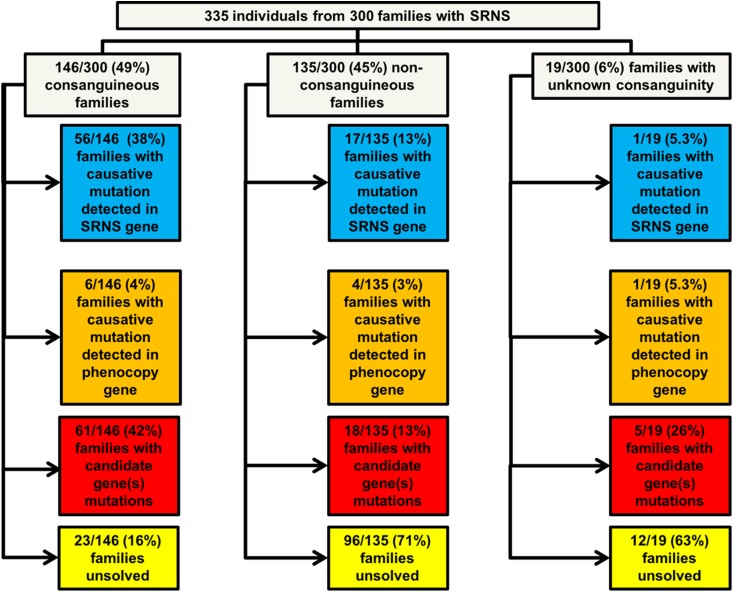
One hundred forty-six of 300 (49%) families were consanguineous, in 56 (38%) of whom we detected a causative mutation in a steroid-resistant nephrotic syndrome gene (Figure 4, Supplemental Table 7). In 56 of 147 families with >100 Mbp of homozygosity on mapping (38%), a causative mutation was detected in a steroid-resistant nephrotic syndrome gene. In contrast, in 17 of 135 (13%) nonconsanguineous families and 18 of 153 (12%) families with <100 Mbp of homozygosity (nonhomozygous) on mapping, a causative mutation was detected in a steroid-resistant nephrotic syndrome gene (Figure 4). The difference in mutation detection between consanguineous and nonconsanguineous families and between homozygous and nonhomozygous families was statistically significant using a chi-squared test (P<0.001) (Figure 4). There was no significant difference in the rate of mutation detection when comparing families with one affected individual versus two affected individuals or ≥3 affected individuals (Figure 4).
Figure 4.
Gene identification in a steroid-resistant nephrotic syndrome gene occurs in a statistically significant greater proportion of homozygous families (homozygosity >100 Mb) when compared to non-homozygous families (homozygosity <100 Mb) and in a statistically significant greater proportion of consanguineous families when compared to non-consanguineous families. Families in whom causative mutations in a known steroid-resistant nephrotic syndrome gene (blue) or a phenocopy gene (orange) were detected, compared with those families in whom no causative mutation was detected (gray). Bars and numbers at end of bars represent total number of families in each category, divided into those families with a causative mutation detected (blue), those families with a causative mutation detected in a phenocopy gene (orange), and those families without a causative mutation detected (gray). Percentages at end of each bar reflect the same three categories. Percentages>10% are rounded to the nearest whole number. Rate of detection of a causative mutation in a steroid-resistant nephrotic syndrome gene did not vary with number of affected individuals per family. Number of affected individuals per family did not have a statistically significant difference between one affected individual per family versus two affected individuals, or between one affected individual and ≥3 individuals. Mutation detection rate in a steroid-resistant nephrotic syndrome gene was significantly higher in those families that were reported clinically as consanguineous or had homozygosity on mapping >100 Mbp than those that were nonconsanguineous or had homozygosity<100 Mbp on mapping (two-sided chi-squared test P<0.001 for each condition). Data of the characteristics of the steroid-resistant nephrotic syndrome cohort compared with the subcohort of those families with a causative mutation detected in a steroid-resistant nephrotic syndrome gene or phenocopy gene are given in Supplemental Table 4. *Statistically significant. SRNS, steroid-resistant nephrotic syndrome.
In 24% of those with additional systemic manifestations in addition to kidney disease, a causative mutation was detected in a steroid-resistant nephrotic syndrome gene, compared with 27% of those with no additional systemic manifestations with a causative mutation detected in a steroid-resistant nephrotic syndrome gene (Supplemental Figure 2, Supplemental Table 8). This difference was not statistically significant.
The most common clinical diagnosis was steroid-resistant nephrotic syndrome in 205 of 300 (68%) (Supplemental Figure 3, Supplemental Table 8). It was the most common clinical diagnosis in those families with a causative mutation identified (48 of 74 families, 65%) (Supplemental Figure 3, Supplemental Table 8).
In 24% of individuals with FSGS on biopsy and in 14% of individuals with diffuse mesangial sclerosis on biopsy, a causative mutation was detected in a steroid-resistant nephrotic syndrome gene (Supplemental Figure 4, Supplemental Table 8). Two hundred twenty-three of 335 (66.6%) individuals had kidney histology data available.
Discussion
Summary and Effect of this Work
To date, this is the largest international cohort in which molecular causes of steroid-resistant nephrotic syndrome were evaluated using whole exome sequencing. Our rate of mutation detection is 25%, consistent with our previous work (8). Recently, in 187 individuals, a causative mutation was detected in one of 53 steroid-resistant nephrotic syndrome genes in 26% of individuals (10).
We detected causative mutations in 20 of 33 known causes of steroid-resistant nephrotic syndrome, with a total of 68 different mutations, 15 of which have not been reported in the literature. To determine the pathogenicity of novel mutations in genes previously described to cause steroid-resistant nephrotic syndrome, we used a strict set of criteria separately for recessive or dominant. Criteria were on the basis of evolutionary conservation, bioinformatic prediction programs of pathogenicity, and allele frequency in healthy control populations (Supplemental Tables 3 and 9).
Before 2014, our lab screened for mutations in NPHS2 and WT1, which may account for lower prevalence in our cohort. NPHS2 and WT1, two of the three most commonly mutated genes in early-onset steroid-resistant nephrotic syndrome, are underrepresented in the presented work, being together responsible for only 3.3% (Table 1) of 300 cases, whereas they were previously reported to be responsible for 15% of cases in 1783 cases (8). When all 1989 families studied in Sadowski et al. (8) and in this study are combined together, mutation rates for NPHS2 and WT1 become more representative of what has been previously published. NPHS2 has a detection rate of 9.3% (185 of 1989) and WT1 has a detection rate of 4.4% (87 of 1989). Mutation rates for NPHS2 were previously 9.9% and for WT1 were 4.8%.
We detected mutations in eight genes that are phenocopies for steroid-resistant nephrotic syndrome, with five novel mutations and five mutations previously reported in the literature. Because these genes may be excluded from panels that target steroid-resistant nephrotic syndrome specifically, these families may have been left without a molecular diagnosis.
Whole exome sequencing allows for the identification of novel genes using homozygosity mapping in consanguineous families and linkage analysis in related individuals. Panels are limited as to how many genes could be evaluated in a given experiment. However, whole exome sequencing allows for the evaluation of all genes, including those that may phenocopy steroid-resistant nephrotic syndrome and provide the opportunity for novel gene discovery.
Therapeutic Implications
Identification of a monogenic cause of steroid-resistant nephrotic syndrome has significant therapeutic implications. (1) In children, treatment often requires prolonged steroid exposure and potentially exposure to multiple immunosuppressant medications. All of these medications carry significant side effect profiles, including growth failure (steroids), bone marrow suppression (mycophenolate mofetil, tacrolimus, azathioprine), kidney dysfunction (tacrolimus), and unacceptable cosmetic effects (cyclosporine), among other side effects. This generates an indication for fast, efficient exome sequencing in order to avoid unfavorable side effects which may be experienced while taking medications that may not provide clinical benefit. (2) Identification of a causative mutation may reveal that a potential therapy is available for some rare single-gene causes of nephrosis. For example, if a mutation in a gene of coenzyme Q10 biosynthesis (COQ2, COQ6, ADCK4, or PDSS2) is detected, treatment with coenzyme Q10 may be indicated (26–28). In the case of the individual with COQ2 mutation, this individual was placed on COQ10 therapy and experienced a sustained remission of nephrosis. (3) Identification of causative mutations in WT1 can also lead to surgical evaluation and intervention to remove streak gonads with potential for malignant transformation (29). (4) Genotype and phenotype correlations in the future may lead to stratification in clinical trials for novel therapeutics. In our study, we identify five families with the p.R1160* mutation in NPHS1. This mutation has been shown to cause congenital nephrotic syndrome; however, in some patients with this mutation, a milder phenotype has been reported (30). (5) Furthermore, identification of mutations in genes that may represent phenocopies of steroid-resistant nephrotic syndrome, such as cystinosis, hyperoxaluria, or Fabry’s disease, can direct therapy. Mutations in these genes would be missed in panel sequencing, because they are not canonic nephrosis genes, but may present with proteinuria, edema, and CKD. The ability to detect mutations in genes that represent phenocopies of nephrosis is a benefit of whole exome sequencing over panel sequencing.
Implications for Kidney Transplant
Many patients progress to ESRD, requiring transplantation and dialysis. Given that approximately 30% of all cases of idiopathic FSGS can recur post-transplant, many centers employ increased immunosuppression before and after transplant to prevent recurrence (31,32). Because monogenic forms of FSGS are unlikely to recur, young children could be spared exposure to aggressive immunosuppression and avoid many of the infectious complications seen in transplantation (10,33). Patients with a monogenic cause of nephrosis identified are younger that those that do not have a causative mutation identified (Figure 3, A and B, Supplemental Table 3), which puts them at greater risk for infections post-transplant, including Epstein–Barr virus and cytomegalovirus. A monogenic diagnosis provides the opportunity to reduce immunosuppression and reduce risk of infectious complication.
Furthermore, many pediatric patients receive a living donor kidney transplant from a close relative, such as a parent, and having a monogenic cause identified, such as COL4A5, may have implications on donor selection. Additionally, in families with INF2 mutations, the parents and family members should be screened for INF2 mutations, because this dominant disease has variable expressivity within families. Should a family member be positive for mutation, this would disqualify them from donation of a kidney for transplantation because they are at risk for developing proteinuria and kidney disease in the future.
Limitations
One limitation to our study is that approximately 70% of families remain undiagnosed. Our lab is currently performing trio analysis, in which both parents and the proband have sequencing performed, which allows for evaluation of nonconsanguineous individuals. We anticipate that this may increase the number of candidate genes identified and lead to future molecular genetic diagnoses.
Cost of Whole Exome Sequencing
Whole exome sequencing has several advantages over panel sequencing. It has the theoretic likelihood of 86% of detecting recessive disease mutations. Whole exome sequencing examines all exons in a genome, whereas panel sequencing typically examines only approximately 30. With falling costs of sequencing, a research exome is approximately $650, which ultimately is more cost effective than panel sequencing because hundreds of panels would be required to cover the whole exome and would not provide the opportunity to identify for novel causes of disease. Once an exome is performed, the data can be revisited as more genes are discovered. With the introduction of trio analysis, nonconsanguineous families can be evaluated for novel genes, potentially allowing for a conclusive monogenic diagnosis in the future.
Disclosures
Friedhelm Hildebrandt has intellectual property licensed to Claritas and is a cofounder of Goldfinch.
Supplementary Material
Acknowledgments
We thank the physicians and the participating families for their contribution. F. Alkandary, Hawally (Kuwait), M. Al Shebadi, and B. Alhermi, Manama (Bahrain); A. Al Swaid, Riyadh (Saudi Arabia), D. Aviles, New Orleans, Louisiana; A. Bakkaloglu, Ankara (Turkey); A. Bagga, New Delhi (India); M. Banks, Denver, Colorado; B. Beck, Cologne (Germany); J. Behunova, Kosice (Slovakia); E. Ben Shalom, Jerusalem (Israel); A. Berdeli, Izmir (Turkey); C. Brix, Hamburg (Germany); J. Camacho, Irvine, California; B. Cavan, Cebu City (Philippines); R. Cohn, Chicago, Illinois; G.S. Ch’ng, Kuala Lumpur (Malaysia); R. Cohen, Jerusalem (Israel); E. Comak, Antalya (Turkey); I. Dursun, Kayseri (Turkey); E. Epstein, Oakland, California; E. Evani and D. Milford, Birmingham (United Kingdom); R. Fine, Los Angeles, California; J. Flynn, Seattle, Washington; G. Giannico, Nashville, Tennessee; K. Gunther, St. Gallen (Switzerland); K. Haffner, Freiburg (Germany); M. Halty, Montevideo (Uruguay); C. Hanna, Minneapolis, Minnesota; M. Hashim, Chennai (India); J. Hernandez, Spokane, Washington; J. Hogan, Philadelphia, Pennsylvania; R. Jenkins, Portland, Oregon. R. Johnson, Denver, Colorado; C. Kempf, Berlin (Germany); W.T. Keng, Kuala Lumpus (Malaysia); S. Kalman, Ankara (Turkey); E. Kamil, Los Angeles, California; D. Katalin, Leipzig (Germany); S. Kerr, Cord Springs, Florida; M. Khemchand, Karachi (Pakistan); K. Kranmiller, Salzburg (Austria); J. Kwinta-Rybicka, Krakow (Poland); E. Lahav, Tel Hashomer (Israel); D. Lewis, Sydney (Australia); K. Lhotta, Feldulrich (Austria); C. Licht and J. Dotsch, Koln (Germany); V. Logan and P. Conlon, Dublin (Ireland); C. Mache, Graz (Austria); M. Mayr, Basel (Switzerland); J. Misselwitz, Jena (Germany); T. Mueller, Vienna (Austria); J. Muhleder and M. Prenninger, Wels (Austria); C. Nailescu, Indianapolis, Indiana; S. Nampoothiri, Kerala (India); S. Narayan, Orange, California; A. Nayir, Istanbul (Turkey); S. Nef, Zurich (Switzerland); T. Neuhaus, Zurich (Switzerland); W. Pietee, Kuala Lumpur (Malaysia); M. Pradhan, Philadelphia, Pennsylvania; A. Ramanathan, Chennai (India); C. Rinat and Y. Frishberg Jerusalem (Israel); I. Roberti, West Orange, New Jersey; R. Roszkowska, Białystok (Poland); J. Saland, New York City, New York; M. Schaeffer, Hannover (Germany); R. Schild, Hamburg (Germany); A. Schulze Everding, Muenster (Germany); W. Seeherunvong, Miami, Florida; P. Senguttvuan and A. Kumar, Chennai (India); J. Siegel-Bartel, Phoenix, Arizona; M. Soran, Adana (Turkey); A. Soylu, Izmir (Turkey); M. Stephan, Munchen (Germany); C. Strehlau, Hannover (Germany); T. Sulakova, Ostrava (Czech Republic); K. Supe-Markovina, Stony Brook, New York; A. Szuminska, Białystok (Poland); R. Tapia, Willmington, Delaware; K. Taranta-Janusz, Białystok (Poland); M. Theault, Minneapolis, Mennesota; Y. Tse, Newcastle upon Tyne (United Kingdom); L. Tranebjaerg, Copenhagen (Denmark); S. Twichell, Burlington, Vermont; M. Waszkiewicz-Stojda, Białystok (Poland); R. Weiss, Valhalla, New York; J. Zeig, Prague (Czech Republic).
F.H. is the William E. Harmon Professor of Pediatrics. This research was supported by grants from the National Institutes of Health to F.H. (DK076683) and J.K.W. (DK007726-31A1). The Yale Center for Mendelian Genomics is funded by U54 HG006504 granted to R.P.L. and by the National Institute of Diabetes and Digestive and Kidney Diseases Intramural Research Program to J.B.K. Funding to W.T. through the American Society of Nephrology Benjamin J. Lipps Research Fellowship Award (FP01014311) and DK007726-31A1. H.Y.G. was supported by the Basic Science Research Program through the National Research Foundation of Korea (2015R1D1A1A01056685). T.H. is supported by a Research Fellowship from the Deutsche Forschungsgemeinschaft (DFG) (HE 7456/1-1). T.J.-S. is supported by the DFG (Jo 1324/1-1). A.J.M. is supported by National Institutes of Health T32-AR053461-04 Postdoctoral Fellow Training Grant. M.N. is supported by the Iinuma-Tsuchiya Foundation for Overseas Research. F.O. is supported by the European Community’s Seventh Framework Programme (FP7/2007-2013) (European Consortium for High-Throughput Research in Rare Kidney Diseases, grant 2012-305608). The Nephrogenetics Laboratory at Hacettepe University was established by the Hacettepe University Infrastructure Project (grant 06A101008). A.V. is supported by the Manton Center for Orphan Diseases Research grant. A.T.v.d.V. is supported by the Postdoctoral Research Fellowship (VE 916/1-1) from the DFG. E.W. is supported by the German National Academy of Sciences Leopoldina (LPDS-2015-07).
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
This article contains supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.04120417/-/DCSupplemental.
References
- 1.Primary nephrotic syndrome in children: Clinical significance of histopathologic variants of minimal change and of diffuse mesangial hypercellularity. A Report of the International Study of Kidney Disease in Children. Kidney Int 20: 765–771, 1981 [DOI] [PubMed] [Google Scholar]
- 2.Smith JM, Stablein DM, Munoz R, Hebert D, McDonald RA: Contributions of the transplant registry: The 2006 annual report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS). Pediatr Transplant 11: 366–373, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Cheong HI, Han HW, Park HW, Ha IS, Han KS, Lee HS, Kim SJ, Choi Y: Early recurrent nephrotic syndrome after renal transplantation in children with focal segmental glomerulosclerosis. Nephrol Dial Transplant 15: 78–81, 2000 [DOI] [PubMed] [Google Scholar]
- 4.Senggutuvan P, Cameron JS, Hartley RB, Rigden S, Chantler C, Haycock G, Williams DG, Ogg C, Koffman G: Recurrence of focal segmental glomerulosclerosis in transplanted kidneys: Analysis of incidence and risk factors in 59 allografts. Pediatr Nephrol 4: 21–28, 1990 [DOI] [PubMed] [Google Scholar]
- 5.Hamiwka LA, Midgley JP, Wade AW, Martz KL, Grisaru S. Outcomes of kidney transplantation in children with nephronophthisis: an analysis of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS) Registry. Pediatr Transplant 12: 878–882, 2008 [DOI] [PubMed] [Google Scholar]
- 6.Lovric S, Ashraf S, Tan W, Hildebrandt F: Genetic testing in steroid-resistant nephrotic syndrome: When and how? Nephrol Dial Transplant 31: 1802–1813, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiggins RC: The spectrum of podocytopathies: A unifying view of glomerular diseases. Kidney Int 71: 1205–1214, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, Engelmann S, Vega-Warner V, Fang H, Halbritter J, Somers MJ, Tan W, Shril S, Fessi I, Lifton RP, Bockenhauer D, El-Desoky S, Kari JA, Zenker M, Kemper MJ, Mueller D, Fathy HM, Soliman NA, Hildebrandt F; SRNS Study Group : A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 26: 1279–1289, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun DA, Schueler M, Halbritter J, Gee HY, Porath JD, Lawson JA, Airik R, Shril S, Allen SJ, Stein D, Al Kindy A, Beck BB, Cengiz N, Moorani KN, Ozaltin F, Hashmi S, Sayer JA, Bockenhauer D, Soliman NA, Otto EA, Lifton RP, Hildebrandt F: Whole exome sequencing identifies causative mutations in the majority of consanguineous or familial cases with childhood-onset increased renal echogenicity. Kidney Int 89: 468–475, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bierzynska A, McCarthy HJ, Soderquest K, Sen ES, Colby E, Ding WY, Nabhan MM, Kerecuk L, Hegde S, Hughes D, Marks S, Feather S, Jones C, Webb NJ, Ognjanovic M, Christian M, Gilbert RD, Sinha MD, Lord GM, Simpson M, Koziell AB, Welsh GI, Saleem MA: Genomic and clinical profiling of a national nephrotic syndrome cohort advocates a precision medicine approach to disease management. Kidney Int 91: 937–947, 2017 [DOI] [PubMed] [Google Scholar]
- 11.Gee HY, Otto EA, Hurd TW, Ashraf S, Chaki M, Cluckey A, Vega-Warner V, Saisawat P, Diaz KA, Fang H, Kohl S, Allen SJ, Airik R, Zhou W, Ramaswami G, Janssen S, Fu C, Innis JL, Weber S, Vester U, Davis EE, Katsanis N, Fathy HM, Jeck N, Klaus G, Nayir A, Rahim KA, Al Attrach I, Al Hassoun I, Ozturk S, Drozdz D, Helmchen U, O’Toole JF, Attanasio M, Lewis RA, Nürnberg G, Nürnberg P, Washburn J, MacDonald J, Innis JW, Levy S, Hildebrandt F: Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int 85: 880–887, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, Tikhonova IR, Bjornson R, Mane SM, Colussi G, Lebel M, Gordon RD, Semmekrot BA, Poujol A, Välimäki MJ, De Ferrari ME, Sanjad SA, Gutkin M, Karet FE, Tucci JR, Stockigt JR, Keppler-Noreuil KM, Porter CC, Anand SK, Whiteford ML, Davis ID, Dewar SB, Bettinelli A, Fadrowski JJ, Belsha CW, Hunley TE, Nelson RD, Trachtman H, Cole TR, Pinsk M, Bockenhauer D, Shenoy M, Vaidyanathan P, Foreman JW, Rasoulpour M, Thameem F, Al-Shahrouri HZ, Radhakrishnan J, Gharavi AG, Goilav B, Lifton RP: Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482: 98–102, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaki M, Airik R, Ghosh AK, Giles RH, Chen R, Slaats GG, Wang H, Hurd TW, Zhou W, Cluckey A, Gee HY, Ramaswami G, Hong C-J, Hamilton BA, Červenka I, Ganji RS, Bryja V, Arts HH, van Reeuwijk J, Oud MM, Letteboer SJ, Roepman R, Husson H, Ibraghimov-Beskrovnaya O, Yasunaga T, Walz G, Eley L, Sayer JA, Schermer B, Liebau MC, Benzing T, Le Corre S, Drummond I, Janssen S, Allen SJ, Natarajan S, O’Toole JF, Attanasio M, Saunier S, Antignac C, Koenekoop RK, Ren H, Lopez I, Nayir A, Stoetzel C, Dollfus H, Massoudi R, Gleeson JG, Andreoli SP, Doherty DG, Lindstrad A, Golzio C, Katsanis N, Pape L, Abboud EB, Al-Rajhi AA, Lewis RA, Omran H, Lee EYHP, Wang S, Sekiguchi JM, Saunders R, Johnson CA, Garner E, Vanselow K, Andersen JS, Shlomai J, Nurnberg G, Nurnberg P, Levy S, Smogorzewska A, Otto EA, Hildebrandt F: Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 150: 533–548, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machuca E, Hummel A, Nevo F, Dantal J, Martinez F, Al-Sabban E, Baudouin V, Abel L, Grünfeld JP, Antignac C: Clinical and epidemiological assessment of steroid-resistant nephrotic syndrome associated with the NPHS2 R229Q variant. Kidney Int 75: 727–735, 2009 [DOI] [PubMed] [Google Scholar]
- 15.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR: A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081, 2009 [DOI] [PubMed] [Google Scholar]
- 17.Schwarz JM, Cooper DN, Schuelke M, Seelow D: MutationTaster2: Mutation prediction for the deep-sequencing age. Nat Methods 11: 361–362, 2014 [DOI] [PubMed] [Google Scholar]
- 18.Hildebrandt F, Heeringa SF, Rüschendorf F, Attanasio M, Nürnberg G, Becker C, Seelow D, Huebner N, Chernin G, Vlangos CN, Zhou W, O’Toole JF, Hoskins BE, Wolf MT, Hinkes BG, Chaib H, Ashraf S, Schoeb DS, Ovunc B, Allen SJ, Vega-Warner V, Wise E, Harville HM, Lyons RH, Washburn J, Macdonald J, Nürnberg P, Otto EA: A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet 5: e1000353, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES: Parametric and nonparametric linkage analysis: A unified multipoint approach. Am J Hum Genet 58: 1347–1363, 1996 [PMC free article] [PubMed] [Google Scholar]
- 20.Strauch K, Fimmers R, Kurz T, Deichmann KA, Wienker TF, Baur MP: Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: Application to mite sensitization. Am J Hum Genet 66: 1945–1957, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A: Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25: 12–13, 2000 [DOI] [PubMed] [Google Scholar]
- 22.Sayer JA, Otto EA, O’Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, Utsch B, Khanna H, Liu Y, Drummond I, Kawakami I, Kusakabe T, Tsuda M, Ma L, Lee H, Larson RG, Allen SJ, Wilkinson CJ, Nigg EA, Shou C, Lillo C, Williams DS, Hoppe B, Kemper MJ, Neuhaus T, Parisi MA, Glass IA, Petry M, Kispert A, Gloy J, Ganner A, Walz G, Zhu X, Goldman D, Nurnberg P, Swaroop A, Leroux MR, Hildebrandt F: The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet 38: 674–681, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup : The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA: From fastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 43: 11.10.1–11.10.33, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P: HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res 37: W593–W599, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montini G, Malaventura C, Salviati L: Early coenzyme Q10 supplementation in primary coenzyme Q10 deficiency. N Engl J Med 358: 2849–2850, 2008 [DOI] [PubMed] [Google Scholar]
- 27.Heeringa SF, Chernin G, Chaki M, Zhou W, Sloan AJ, Ji Z, Xie LX, Salviati L, Hurd TW, Vega-Warner V, Killen PD, Raphael Y, Ashraf S, Ovunc B, Schoeb DS, McLaughlin HM, Airik R, Vlangos CN, Gbadegesin R, Hinkes B, Saisawat P, Trevisson E, Doimo M, Casarin A, Pertegato V, Giorgi G, Prokisch H, Rötig A, Nürnberg G, Becker C, Wang S, Ozaltin F, Topaloglu R, Bakkaloglu A, Bakkaloglu SA, Müller D, Beissert A, Mir S, Berdeli A, Varpizen S, Zenker M, Matejas V, Santos-Ocaña C, Navas P, Kusakabe T, Kispert A, Akman S, Soliman NA, Krick S, Mundel P, Reiser J, Nürnberg P, Clarke CF, Wiggins RC, Faul C, Hildebrandt F: COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest 121: 2013–2024, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atmaca M, Gulhan B, Korkmaz E, Inozu M, Soylemezoglu O, Candan C, Bayazıt AK, Elmacı AM, Parmaksiz G, Duzova A, Besbas N, Topaloglu R, Ozaltin F: Follow-up results of patients with ADCK4 mutations and the efficacy of CoQ10 treatment. Pediatr Nephrol 32: 1369–1375, 2017 [DOI] [PubMed] [Google Scholar]
- 29.Chernin G, Vega-Warner V, Schoeb DS, Heeringa SF, Ovunc B, Saisawat P, Cleper R, Ozaltin F, Hildebrandt F; Members of the GPN Study Group : Genotype/phenotype correlation in nephrotic syndrome caused by WT1 mutations. Clin J Am Soc Nephrol 5: 1655–1662, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koziell A, Grech V, Hussain S, Lee G, Lenkkeri U, Tryggvason K, Scambler P: Genotype/phenotype correlations of NPHS1 and NPHS2 mutations in nephrotic syndrome advocate a functional inter-relationship in glomerular filtration. Hum Mol Genet 11: 379–388, 2002 [DOI] [PubMed] [Google Scholar]
- 31.Sener A, Bella AJ, Nguan C, Luke PP, House AA: Focal segmental glomerular sclerosis in renal transplant recipients: Predicting early disease recurrence may prolong allograft function. Clin Transplant 23: 96–100, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez E, Ettenger R, Rianthavorn P, Tsai E, Malekzadeh M: Preemptive plasmapheresis and recurrence of focal segmental glomerulosclerosis in pediatric renal transplantation. Pediatr Transplant 15: 495–501, 2011 [DOI] [PubMed] [Google Scholar]
- 33.Fornoni A, Sageshima J, Wei C, Merscher-Gomez S, Aguillon-Prada R, Jauregui AN, Li J, Mattiazzi A, Ciancio G, Chen L, Zilleruelo G, Abitbol C, Chandar J, Seeherunvong W, Ricordi C, Ikehata M, Rastaldi MP, Reiser J, Burke GW 3rd : Rituximab targets podocytes in recurrent focal segmental glomerulosclerosis. Sci Transl Med 3: 85ra46, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.