Abstract
PLAZA (https://bioinformatics.psb.ugent.be/plaza) is a plant-oriented online resource for comparative, evolutionary and functional genomics. The PLAZA platform consists of multiple independent instances focusing on different plant clades, while also providing access to a consistent set of reference species. Each PLAZA instance contains structural and functional gene annotations, gene family data and phylogenetic trees and detailed gene colinearity information. A user-friendly web interface makes the necessary tools and visualizations accessible, specific for each data type. Here we present PLAZA 4.0, the latest iteration of the PLAZA framework. This version consists of two new instances (Dicots 4.0 and Monocots 4.0) providing a large increase in newly available species, and offers access to updated and newly implemented tools and visualizations, helping users with the ever-increasing demands for complex and in-depth analyzes. The total number of species across both instances nearly doubles from 37 species in PLAZA 3.0 to 71 species in PLAZA 4.0, with a much broader coverage of crop species (e.g. wheat, palm oil) and species of evolutionary interest (e.g. spruce, Marchantia). The new PLAZA instances can also be accessed by a programming interface through a RESTful web service, thus allowing bioinformaticians to optimally leverage the power of the PLAZA platform.
INTRODUCTION
The wealth of data produced by the various sequencing consortia is not utilized to its full potential without the application of in-depth functional genomics, bolstered by robust comparative genomics. Indeed, while the price of sequencing a single genome has dropped dramatically these past few years (1), repeating even a fraction of all experiments performed on model organisms would represent an exorbitant cost. It is therefore imperative that platforms are developed that offer the necessary tools to translate and transfer knowledge from well-studied model organisms to species of economic, ecological and evolutionary interest. Within the plant kingdom a variety of such platforms is available, each with a different focus, such as PLAZA (2), Phytozome (3), Gramene (4) and Ensembl Plants (5).
In this manuscript we present PLAZA 4.0 (available at https://bioinformatics.psb.ugent.be/plaza), the most recent version of the PLAZA platform for comparative, evolutionary and functional genomics. This latest version offers access to a total of 71 species (Supplementary Table S1), for each of which we computed functional and homology information based on the available structural annotations. The PLAZA platform functions as a web-based information and analysis portal to study both model and crop plant organisms through the different data types, analysis tools and visualizations it offers.
OVERVIEW PLANT GENOMES
Driven by the ever-decreasing cost and expanding throughput of new genome sequencing technologies, many new plant genomes have been sequenced during the last years, ranging from small to gigantic, repeat-rich or polyploid genomes (6). This acceleration resulted in a total of 157 available plant genomes of varying quality with regards to both assembly and annotation, covering a wide range of orders and families within the green plant lineage. We chose to incorporate a selection of species, based on three distinct criteria: the phylogenetic clade, the coverage of taxa per clade and the quality of the genome assembly. In total, 71 genomes were retained and distributed over two PLAZA 4.0 instances focusing on dicots and monocots, respectively (see Supplementary Table S1 for data sources and version information, and Supplementary Method S1 for references used to build the instance species trees).
Shared between both PLAZA instances are 13 reference species which largely exhibit the following characteristics: high-quality genome for a model species of a specific phylogenetic clade (e.g. Oryza sativa), large amount of functional data (e.g. Arabidopsis thaliana), unique phylogenetic location and/or use as outgroup (e.g. Physcomitrella patens and Chlamydomonas reinhardtii). The reference species that we are using within both PLAZA instances also include species that were not present in previous PLAZA versions, but which are deemed crucial as outgroups to the flowering plants. For example, Amborella trichopoda was only present in the dicot instance of PLAZA 3.0 but is a reference species in both PLAZA 4.0 instances due to its unique place in the phylogeny of the Angiosperms. With the addition of Picea abies, a representative of the Gymnosperm clade has been included, and Marchantia polymorpha strengthens our understanding of the origins of seed plants as it is part of the same extant clade as Physcomitrella patens. Since dedicated resources focusing on unicellular photosynthetic eukaryotes are available elsewhere (7,8), we decided to only incorporate a limited set of green algae to be used as outgroup for the new PLAZA 4.0 instances.
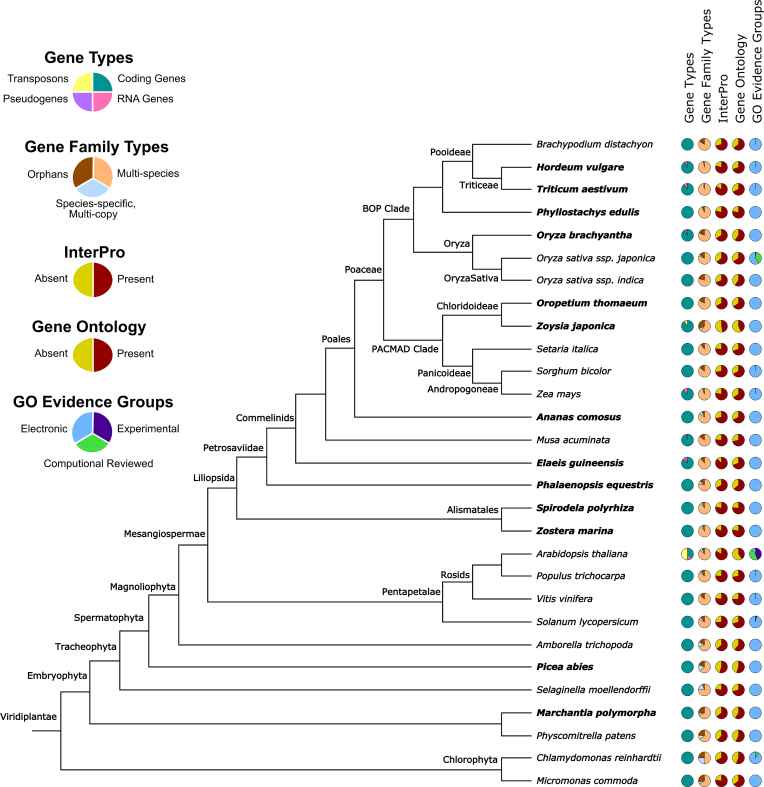
Both the PLAZA 4.0 Monocot and Dicot instances contain significantly more species compared to their PLAZA 3.0 counterparts, having 29 and 55 genomes, respectively. For the Dicot instance the largest increase is observed within the Asterid clade, which now contains nine species covering six different families. The two other major clades within the Eudicots are the Malvids and the Fabids. The coverage of the former has increased only moderately (from 10 to 13 species), while the latter nearly doubled in size compared to PLAZA 3.0 (from 11 to 20 species). For the Monocot instance a more gradual expansion of the different clades can be seen (Figure 1) with the total number of Monocots increasing from 8 to 18 species. PLAZA 4.0 now covers a larger number of different Monocot families (12 families versus 5 families in 3.0), including the introduction of early branching clades such as the Alismatales (Zostera marina and Spirodela polyrhiza), the Asparagales (Phalaenopsis equestris) and the Arecales (Elaeis guineensis).
Figure 1.
Data content overview Monocot instance PLAZA 4.0. Species in bold are newly included species compared to previous PLAZA instances. Orphan gene families are single-copy species-specific genes. GO evidence groups are based on the following GO evidence types: Experimental (EXP, IDA, IPI, IMP, IGI, IEP), Computational Reviewed (ISS, ISO, ISA, ISM, IGC, IBA, IBD, IKR, IRD, RCA) and Electronic (TAS, NAS, IC, ND, IEA, NR).
To ensure maximum compatibility with other platforms, the PLAZA 4.0 platform now aims to use the original gene identifiers as provided by the genome sequencing projects. In previous PLAZA versions custom identifiers were often created in order to have a clean and consistent nomenclature of genes, in line with the TAIR gene identifiers. This approach has however become untenable: keeping stable gene identifiers across multiple annotation versions is a priority for many genome annotation projects, but requires considerable effort and is not easily duplicated for the large amount of organisms within the PLAZA platform. Therefore, we strived to retain the original gene identifiers wherever possible (see Supplementary Table S1 for exceptions). All search functions, as well as the PLAZA workbench, support both the original as well as newly generated gene identifiers.
NEW FEATURES TO IMPROVE COMPARATIVE GENOMICS DATA EXPLORATION
New and improved visualizations
Offering a web-based platform, which holds large and complex data, to end-users often entails multi-tiered solutions: while some users prefer simply browsing column-based information, others require attractive and informative visualizations to reduce the complexity of the data to make them more easily interpretable. Therefore, multiple custom visualizations and interactive tools are under continuous development in the PLAZA platform. With the major browsers moving away from plugins such as Java, Silverlight and Flash, it has become imperative to replace the tools that depended on these plugins with newly developed state-of-the-art equivalents. Furthermore, some of the tools present in previous PLAZA versions relied on static images with very few possibilities for interaction. These were thus also replaced in order to further allow the end-user to explore the data in a user-friendly and efficient way.
Phylogeny is one of the core concepts within the PLAZA platform: from studying orthologous relationships to the transfer of functional annotation, many tools and analyzes depend on the visualization and interpretation of phylogenetic trees. This feature has been enhanced by mapping additional information onto phylogenetic trees (such as gene structure information, protein domains or gene duplication information), helping the downstream analysis of the associated gene families. The previous PLAZA versions relied on the externally developed software tool Archaeopteryx (previously ATV) (9) to display these phylogenetic trees. However, as Archaeopteryx is a Java applet-based solution, we developed PhyD3 (10), an easily extensible JavaScript based alternative that can be used to add a variety of data charts to phylogenetic trees. PhyD3 can be used to visualize species trees as well as gene family trees.
A tree-based visualization made using PhyD3 was developed to generate a data overview of the content within the PLAZA platform (Figure 1 and Supplementary Figure S1). This overview offers an intuitive way to compare the annotations, both structural and functional, between the various species present in a PLAZA instance. For example, by exploring these data overviews it quickly becomes quite clear that only few genome projects include the annotation of non-coding genes, and if they do it is mostly restricted to transposable elements. Similarly, this view offers a clear summary of the available functional annotations per species, enhanced with evidence information for Gene Ontology (GO) annotations over three categories (experimental, computational reviewed and electronic annotations).
Whereas the visualization of multiple sequence alignments (MSAs) was previously performed using the Java program JalView (11), a JavaScript implementation present in the BioJS registry (12) is now used to explore MSAs (13). Although less extensive with regards to options and associated tools when compared to JalView, this JavaScript implementation still offers the necessary basics which are expected of an MSA viewer: sorting of sequences, filtering of both columns and sequences based on various criteria, selecting different coloring schemes, and exporting the MSA in various output formats.
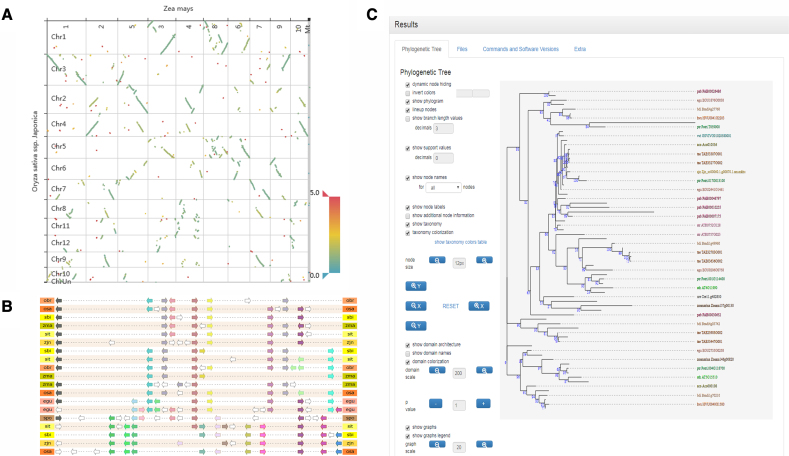
Gene and genome colinearity is another major data type within the PLAZA platform. Colinear regions, describing conserved gene content and order (both within and between species) are instrumental in the study of genome evolution (14) and can also be used to identify orthologs showing conserved genome organization. Within the PLAZA platform, colinearity is visualized on three different levels: between species, between chromosomes, and between chromosomal segments. For the first two levels the most-used exploratory visualization is the WGDotplot tool (15). Originally only a static image was provided with more interactivity provided in subsequent updates by the introduction of a Java applet-based solution (16). Now, a custom JavaScript replacement has been implemented, offering the same set of options while being future-proof and less memory intensive (Figure 2A). The colinearity between chromosomal segments requires more advanced data visualizations, as colinear regions between multiple species can span multiple segments. A set of colinear segments is indicated with the term ‘Multiplicon’, and associated with this data type the MultipliconViewer tool is implemented. A previously implemented static image-based visualization has now been replaced by a more flexible and interactive JavaScript-based viewer. The new MultipliconViewer offers adaptive filtering (allowing end-users to view only the segments of the species of interest), custom sorting (rearranging the order of the colinear regions within the view), as well as much more detailed and customizable information when selecting genes within the colinear regions (Figure 2B). In PLAZA 4.0 colinear regions are now also searchable through the Colinear Region Finder tool, making it now possible to look for multiplicons that have complex and adaptable strictness requirements (e.g. regions that are present in all dicots, duplicated in A. thaliana but not duplicated in Brassica rapa).
Figure 2.
New tools and data visualizations. From top to bottom and left to right: (A) JavaScript WGDotPlot, (B) Multiplicon Plot, (C) Interactive Phylogenetics Module. The WGDotPlot shows the inter-species colinear regions between Oryza sativa ssp. Japonica and Zea mays. The Multiplicon Plot shows highly conserved colinearity across the Angiosperms (PLAZA 4.0 Monocot instance). The Interactive Phylogenetics Module was run using family HOM04M002831 as seed with a custom selection of species.
Compared to colinearity, synteny is a less strict alternative where only the conservation of gene content is considered (14). By using the SyntenyPlot visualization, genomic adaptations between gene family members, such as gene insertions, gene inversions and gene transpositions, are easily investigated. The new SyntenyPlot is based on the same codebase as the MultipliconViewer and as such offers the same functionality: filtering, sorting and showing additional gene content information (Supplementary Figure S2). To cope with large gene families, data is presented to the end-user in a paginated way within the SyntenyPlot.
In order to make the website fully compatible with mobile devices, the GenomeView genome browser has been replaced with IGV.js genome browser (https://igv.org/), which is a JavaScript implementation of the original IGV genome browser (17).
Adaptations to the PLAZA workbench
The PLAZA workbench allows users to perform analyses on sets of genes, without requiring any programming experience. Since its inception in the first PLAZA version (15), multiple improvements have been made with regards to its functionality and performance (16). In PLAZA 4.0, we introduce the feature to share workbench experiments between end-users: a single user can now create a workbench experiment and share this with other users. This prevents users working on the same dataset from having to maintain their own workbench experiments and having to synchronize between different researchers
Interactive phylogenetics module
For every gene family present in the PLAZA platform, a maximum likelihood phylogenetic tree is computed. Whereas PLAZA 4.0 Dicots and Monocots contain 24 580 and 19 125 pre-computed phylogenetic trees, respectively, one of the drawbacks of a fully-automated build procedure is that certain limitations have to be set with regards to the computation of MSAs and phylogenetic trees. For very large gene families (>1000 genes) the PLAZA workflow does not compute the phylogenetic trees due to potential low quality MSAs which do not result in reliable phylogenetic trees.
To overcome this limitation, a new tool was developed that allows the user to create a custom MSA and phylogenetic tree by using the PLAZA platform as reference. This interactive phylogenetics module offers a versatile tool for customized gene orthology analysis, by giving the user the ability to select which species or genes to include in the MSA and tree. Gene families for which a tree is computed often contain genes from species that the end-user is not interested in, and gene families which are too large need gene selection and filtering to be interpretable. Furthermore, extra homologous DNA or protein sequences can be added, enabling the inclusion of genes from species not present in the PLAZA platform.
The Interactive Phylogenetics Module can either start from a single gene or from a gene family, which is used as seed to define the gene set for which the MSA and phylogenetic tree will be constructed. Genes can be added to the gene set in multiple ways: (i) single-gene selection based on the BLAST-hits of the query gene or gene family members, (ii) species-based selection based on the BLAST-hits of the query gene or gene family members, (iii) external DNA or protein sequence(s). After the gene selection, the user can proceed by defining the program settings. While the standard PLAZA routine for tree construction consist of MSA creation by MUSCLE (18) followed by MSA filtering and trimming, and phylogenetic tree construction using FastTree (19), the Interactive Phylogenetics Module allows for different processing options. The algorithm used for the generation of the MSA can be either MUSCLE or MAFFT (20), the latter using automatic method detection. Next, the user can choose the MSA editing mode: no editing, removal of lowly conserved positions by trimming the sequences, filtering of partial sequences or both trimming and filtering. Finally, the phylogenetic tree construction method has to be defined, which can be either the approximate maximum likelihood method FastTree, or the maximum likelihood methods PhyML (21), RaXML (22) or IQ-TREE (23). The latter will perform a test to detect the best fitting protein model (24) from the following widely used models: JTT, LG, WAG, Blosum62, VT and Dayhoff. A thousand rounds of ultra-parametric bootstrapping are run in IQ-TREE and the FreeRate model is used for rate heterogeneity across sites. All other methods use the WAG protein model in combination with 100 bootstraps and gamma approximation for modeling rate heterogeneity across sites. The resulting tree is then visualized using the PhyD3 program (Figure 2C).
Application programming interface (API)
The PLAZA platform currently performs multiple roles: (i) a starting-point for single-gene analysis, (ii) a reference for orthologous relationships and (iii) a tool to study genome evolution. In addition PLAZA also functions as a reference data warehouse: multiple papers cite PLAZA as the platform where they retrieved the structural and/or functional annotation of entire species to be used in other large-scale analyzes (25,26). This is made possible because the PLAZA platform offers downloads for both the input data and processed data in standardized formats. However, this approach is cumbersome and sub-optimal for other online tools which would like to make use of the PLAZA data in a more interactive way, e.g. workflow systems such as Galaxy (27). To accommodate this growing need, the PLAZA platform now offers access to multiple data types through a newly developed Application Programming Interface (API).
This RESTful API is available per PLAZA instance (e.g. Dicots 4.0 or Monocots 4.0). Therefore, the end-user has to decide first which PLAZA instance to query prior to performing the API calls. The API can be accessed either after authentication or anonymously. The former requires an account at the workbench of the PLAZA instance being targeted, and this information is used to obtain a JSON Web Token (JWT). This token is subsequently passed to the server during the follow-up API calls to ensure authentication without having to resend the username and password with each API call, thus limiting the risk of eavesdropping. The use of JWT allows for granular filtering of users, as well as for the throttling of API calls to prevent overload of the server. Anonymous access to the API is possible by requesting a token without providing authentication information. The user identifier within this token is however shared between all anonymous users and thus all these requests are subject to server-side throttling of API requests. Users can easily circumvent this by creating a free account in the workbench of the appropriate PLAZA instance.
All major data types from the PLAZA platform can be retrieved using the API, on a gene-by-gene basis or by providing a set of gene identifiers as input: structural annotations, gene family information, functional annotations (Gene Ontology and InterPro), orthology information, etc. Apart from support for data-retrieval methods, some types of analysis methods such as GO enrichment are also provided (Supplementary Figures S3 and 4).
The PLAZA API, like the rest of the platform, is under continuous development. Therefore, the offered API calls will be extended in the future. An up-to-date overview of the PLAZA API calls, as well as example routines in various scripting languages, can be found in the online PLAZA documentation (https://bioinformatics.psb.ugent.be/plaza/documentation).
BUILD PROCEDURE
The general workflow of the PLAZA build procedure, as described in (2,15,16) has not been significantly altered: protein coding genes are compared against each other, subsequently clustered into gene families, and used to delineate colinear regions between and within genomes by application of the i-ADHoRe algorithm (28) (Supplementary Method S2). MSAs and phylogenetic trees are constructed for the gene families. The trees, the subfamily information, the colinearity information and the protein alignment scores are then used to infer orthologous relationships using an integrative approach. This data is subsequently used to transfer functional annotations between orthologs, from model species to crop species.
Some programs within the workflow have been replaced in favor of alternatives offering better performance or efficiency: NCBI BLAST (29) has been replaced by DIAMOND (30), while OrthoMCL (31) has been replaced by OrthoFinder (32). The replacement of NCBI BLAST was decided upon due to the massive computational performance gains introduced by the DIAMOND program, while maintaining sensitivity and specificity comparable to NCBI BLAST. We used the ‘–more-sensitive’ parameter in order to retain the highest possible accuracy, together with the 1e-5 e-value used in previous PLAZA builds (Supplementary Method S2).
CONCLUSION
The PLAZA 4.0 update offers a large increase in the number of available species covering a much broader range of plant families, targeting organisms that are of commercial, ecological or academic interest. The reworked and redesigned visualizations offer an improved user experience by no longer relying on external web browser plugins. With the addition of the new Interactive Phylogenetics Module, the PLAZA platform now also offers a solution to resolve the phylogeny of complex and large gene families. Finally, the newly developed API creates novel opportunities for bioinformaticians to retrieve data from the PLAZA platform in an interactive way.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to express gratitude to the various genome sequencing project managers who have graciously allowed their data to be used within the PLAZA platform, even when the genome itself had not been published yet.
Author contributions: M.V.B. and T.D. parsed genome data. M.V.B. updated and automated the PLAZA pipeline. T.D. helped with the development of the API. L.K. developed the JavaScript WGDotPlot. E.V. helped with the development of the Interactive Phylogenetics Module. M.V.B., E.V. and K.V. wrote the manuscript. A.B., F.C. and Y.V.d.P. played a managing role.
All authors read and approved the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
ELIXIR-EXCELERATE (European Commission within the Research Infrastructures program of Horizon 2020) [676559 to F.C.]; Research Foundation-Flanders (FWO) and Hermesfonds for ELIXIR Belgium [AH.2017.051,IO 17001306 to T.D.]. Funding for open access charge: VIB; Ghent University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Christensen K.D., Dukhovny D., Siebert U., Green R.C.. Assessing the costs and cost-effectiveness of genomic sequencing. J. Pers. Med. 2015; 5:470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Proost S., Van Bel M., Vaneechoutte D., Van de Peer Y., Inze D., Mueller-Roeber B., Vandepoele K.. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 2015; 43:D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N. et al. . Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gupta P., Naithani S., Tello-Ruiz M.K., Chougule K., D’Eustachio P., Fabregat A., Jiao Y., Keays M., Lee Y.K., Kumari S. et al. . Gramene Database: navigating plant comparative genomics resources. Curr. Plant Biol. 2016; 7–8:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bolser D.M., Staines D.M., Perry E., Kersey P.J.. Ensembl Plants: integrating tools for visualizing, mining, and analyzing plant genomic data. Methods Mol. Biol. 2017; 1533:1–31. [DOI] [PubMed] [Google Scholar]
- 6. Veeckman E., Ruttink T., Vandepoele K.. Are we there yet? Reliably estimating the completeness of plant genome sequences. Plant Cell. 2016; 28:1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vandepoele K., Van Bel M., Richard G., Van Landeghem S., Verhelst B., Moreau H., Van de Peer Y., Grimsley N., Piganeau G.. pico-PLAZA, a genome database of microbial photosynthetic eukaryotes. Environ. Microbiol. 2013; 15:2147–2153. [DOI] [PubMed] [Google Scholar]
- 8. Kersey P.J., Allen J.E., Armean I., Boddu S., Bolt B.J., Carvalho-Silva D., Christensen M., Davis P., Falin L.J., Grabmueller C. et al. . Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016; 44:D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han M.V., Zmasek C.M.. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009; 10:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kreft L., Botzki A., Coppens F., Vandepoele K., Van Bel M.. PhyD3: a phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics. 2017; 33:2946–2947. [DOI] [PubMed] [Google Scholar]
- 11. Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J.. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009; 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gomez J., Garcia L.J., Salazar G.A., Villaveces J., Gore S., Garcia A., Martin M.J., Launay G., Alcantara R., Del-Toro N. et al. . BioJS: an open source JavaScript framework for biological data visualization. Bioinformatics. 2013; 29:1103–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yachdav G., Wilzbach S., Rauscher B., Sheridan R., Sillitoe I., Procter J., Lewis S.E., Rost B., Goldberg T.. MSAViewer: interactive JavaScript visualization of multiple sequence alignments. Bioinformatics. 2016; 32:3501–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang H., Bowers J.E., Wang X., Ming R., Alam M., Paterson A.H.. Synteny and collinearity in plant genomes. Science. 2008; 320:486–488. [DOI] [PubMed] [Google Scholar]
- 15. Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K.. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009; 21:3718–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Bel M., Proost S., Wischnitzki E., Movahedi S., Scheerlinck C., Van de Peer Y., Vandepoele K.. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 2012; 158:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thorvaldsdottir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Price M.N., Dehal P.S., Arkin A.P.. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Eevol. 2009; 26:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katoh K., Misawa K., Kuma K., Miyata T.. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002; 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guindon S., Delsuc F., Dufayard J.F., Gascuel O.. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 2009; 537:113–137. [DOI] [PubMed] [Google Scholar]
- 22. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015; 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S.. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017; 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castro P.H., Lilay G.H., Munoz-Merida A., Schjoerring J.K., Azevedo H., Assuncao A.G.L.. Phylogenetic analysis of F-bZIP transcription factors indicates conservation of the zinc deficiency response across land plants. Sci. Rep. 2017; 7:3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tohge T., Watanabe M., Hoefgen R., Fernie A.R.. The evolution of phenylpropanoid metabolism in the green lineage. Crit. Rev. Biochem. Mol. Biol. 2013; 48:123–152. [DOI] [PubMed] [Google Scholar]
- 27. Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Eberhard C. et al. . The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016; 44:W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Proost S., Fostier J., De Witte D., Dhoedt B., Demeester P., Van de Peer Y., Vandepoele K.. i-ADHoRe 3.0–fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 2012; 40:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 30. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 31. Li L., Stoeckert C.J. Jr, Roos D.S.. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003; 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emms D.M., Kelly S.. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015; 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.