Abstract
We introduce the marine databases; MarRef, MarDB and MarCat (https://mmp.sfb.uit.no/databases/), which are publicly available resources that promote marine research and innovation. These data resources, which have been implemented in the Marine Metagenomics Portal (MMP) (https://mmp.sfb.uit.no/), are collections of richly annotated and manually curated contextual (metadata) and sequence databases representing three tiers of accuracy. While MarRef is a database for completely sequenced marine prokaryotic genomes, which represent a marine prokaryote reference genome database, MarDB includes all incomplete sequenced prokaryotic genomes regardless level of completeness. The last database, MarCat, represents a gene (protein) catalog of uncultivable (and cultivable) marine genes and proteins derived from marine metagenomics samples. The first versions of MarRef and MarDB contain 612 and 3726 records, respectively. Each record is built up of 106 metadata fields including attributes for sampling, sequencing, assembly and annotation in addition to the organism and taxonomic information. Currently, MarCat contains 1227 records with 55 metadata fields. Ontologies and controlled vocabularies are used in the contextual databases to enhance consistency. The user-friendly web interface lets the visitors browse, filter and search in the contextual databases and perform BLAST searches against the corresponding sequence databases. All contextual and sequence databases are freely accessible and downloadable from https://s1.sfb.uit.no/public/mar/.
INTRODUCTION
Microorganisms are ubiquitous in the marine environment, where they play key roles in many global and local biogeochemical processes such as nutrient recycling (1). These microorganisms and the communities they form, drive and respond to changes in the environment and alterations in the marine environment (2). With an estimated 104 to 106 cells per milliliter seawater and totally over 1029 bacterial cells in open sea, the marine microorganisms provide the grounds for immense genetic diversity (3).
Since the first complete bacterial genome published in 1995 (4), the number of sequenced microbial genomes has increased dramatically. Currently, more than 103 000 prokaryotic genomes are available in the National Center for Biotechnology Information (NCBI) Genome microbial database (https://www.ncbi.nlm.nih.gov/genome/microbes/). Originally sequencing efforts were prioritized to study cultured microbes. However, it is well established that the vast majority of bacterial and archaeal taxa remain uncultivated in vitro (5). Recently, cultivation-independent methods such as single cell genomics and genomes reconstructed from metagenomic deep sequencing, have begun to yield complete or near-complete genomes from many novel lineages (5–7). Metagenomics, the study of genetic material recovered directly from environmental samples, is a powerful tool for surveying the diversity of marine microbes, which are important for the study of marine sciences. Prominent examples of metagenomics studies in the marine field include the Sorcerer II expeditions (8), Malaspina expedition (9), Global Ocean Sampling (GOS) campaign (10) and Tara Oceans expedition (11). Most of these data as well as other marine metagenomic data are stored in publicly available metagenomic databases such as iMicrobe (https://www.imicrobe.us/), Viral Informatics Resource for Metagenome Exploration (VIROME) (12), EBI metagenomics (13), Integrated Microbial Genomes and Microbiomes (IMG/M) (14) and Metagenomics Rapid Annotation using Subsystem Technology (MG-RAST) (15). Reference sequence databases with comprehensive metadata are essential for analyzing and interpreting of marine metagenomic data (16,17). There are several general microbial databases e.g. Prokaryotic RefSeq Genomes (18), Genomes OnLine Database (GOLD) (19), Pathosystems Resource Integration Center (PATRIC) (20) and MicroScope (21), which contains marine microbial genomes. Even though the Microbial Ecological Genomics Database (MegDB), available at the Megx.net portal, includes marine bacterial, archaeal and phage genomes and metagenomes, it is mainly a georeference database which provides less metadata besides the geolocation information of the samples (22).
Up to now, no dedicated sequence data resources exist for the marine metagenomics domain (17), which not only hamper the utilization of the vast genetic resources for biotechnology research and innovation (e.g. bioprospecting), but also impede the development of sustainable tools and resources aimed at environmental monitoring, monitoring of fish and shellfish pathogens and development of sustainable feed for marine aquaculture.
Since all research and innovation is based on comparison to existing knowledge and information, the lack of unified formats, controlled vocabularies (CV) and ontologies (formal specifications of the terms) make it difficult not only to identify records in databases but also to compare data within and/or between different databases. Therefore, sustainable and highly accurate data resources that are easy to access, browse and retrieve data from, are vital for performing high class and beyond the state of art research and innovation.
Here, we introduced the contextual and sequence MAR databases: MarRef, MarDB and MarCat, with manually curated metadata including attributes for sampling, sequencing, assembly and annotation in addition to the organism and taxonomic information and their corresponding nucleotide and protein sequences.
OVERVIEW OF THE RESOURCES
Definition of marine microbial biome
To define a ‘marine microbial biome’ or a ‘marine microorganism’ is not straightforward since there are many habitats, which are on the borderline between marine and terrestrial ecosystems, such as sandy shores and near river deltas. We have chosen to define a ‘marine microbial biome’ as ‘An aquatic microbial biome comprises of microbial communities from open oceans, coastal and protected habitats up to the high-water mark with salinity from 0.5 ppt (parts per thousand) as in estuaries (brackish water) environments to above 100 ppt as in sea ice brine. The biome also includes marine microbial communities obtained from marine species associated with these habitats’.
Additionally, we accept soil samples from sandy shores, intertidal zones, salt marshes (coastal salt marshes or tidal marshes), mudflats and estuaries, in addition to habitats such as seawater saltern, sea ice brines, black smokers (hydrothermal vents) where the salinity can be extremely high or low compared to the seawater. Microorganisms and microbiomes associated with marine species, as defined by the World Register of Marine Species (WoRMS) have also been defined as marine (23). This includes microorganisms associated with or causing diseases in marine animals and plants such as corals, shellfish, fish, macroalgae and seagrass.
Short description of MarRef, MarDB and MarCat
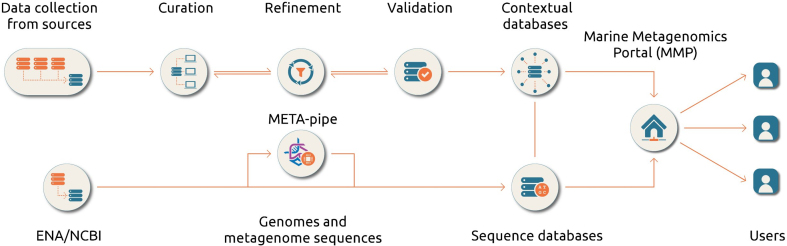
The construction of the marine contextual databases and their corresponding sequence databases (BLAST databases) are shown in Figure 1. Each genome or metagenome assigned to a ‘marine microbial biome’, according to our definition, is included in the databases.
Figure 1.
General and simplified procedures for construction of the MAR databases. The top part represents the flow of contextual data records from its collection to implementation on the web server. The bottom part illustrates how sequence data becomes implemented and processed. Only metagenomic sequences in relation with MarCat has been processed using META-pipe for the first release.
The MarRef, MarDB and MarCat sequence databases are based on the non-redundant genome and metagenome datasets obtained from ENA (European Nucleotide Archive, http://www.ebi.ac.uk/ena) and NCBI (https://www.ncbi.nlm.nih.gov/). While MarRef is a database for completely sequenced marine prokaryotic genomes, MarDB includes all in-complete sequenced marine prokaryotic genomes regardless the level of completeness. MarCat represents a gene (protein) catalog of predicted marine genes derived from marine metagenomic samples. Metagenomic sequences were obtained from ENA and their corresponding gene and protein annotation unique to each sample was generated using META-pipe, a pipeline for taxonomic classification and functional annotation of metagenomic sample (arXiv:1604.04103). The corresponding contextual databases support the international community-driven standards of the Genomics Standards Consortium (http://gensc.org/) and are fully compliant with its recommendations for minimum information about any (x) sequence (MIxS) standards. These databases also include the proposed standards for provenance of analysis proposed by the ELIXIR EXCELERATE marine metagenomics community (24).
CONTEXTUAL DATABASES
Data collection
The MarRef, MarDB and MarCat contextual databases are built by compiling data from a number of publicly available sequence, taxonomy and literature databases in a semi-automatic fashion. Other databases or resources such as bacterial diversity and culture collections databases, web mapping services and ontology databases were used extensively for curation of metadata. Resources used in the curation of the marine databases are shown in Table 1.
Table 1. Public data resources utilized for the construction of MarRef, MarDB and MarCat.
| Type | Database | URL |
|---|---|---|
| Sequence databases | ENA, European Nucleotide Archive | ebi.ac.uk/ena |
| UniProt, Universal Protein Resource | uniprot.org | |
| NCBI, National Center for Biotechnology Information | ncbi.nlm.nih.gov | |
| Contextual databases | PATRIC, Pathosystems Resource Integration Center | patricbrc.org |
| GOLD, Genomes OnLine Database | gold.jgi.doe.gov | |
| Taxonomic databases | SILVA, SILVA high quality ribosomal RNA database | arb-silva.de |
| NCBI Taxonomy browser | ncbi.nlm.nih.gov/taxonomy | |
| Bacterial diversity metadatabases | BacDive, Bacterial Diversity Metadatabase | bacdive.dsmz.de |
| Culture collection databases | DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH | dsmz.de |
| ATCC, American Type Culture Collection | atcc.org | |
| Marine organisms database | WoRMS, World Register of Marine Species | marinespecies.org |
| Web mapping service | Google maps | maps.google.com |
| Literature databases | Europe PMC, Europe PubMed Central | europepmc.org |
| PubMed | ncbi.nlm.nih.gov/pubmed | |
| doi, Digital Object Identifier System | doi.org | |
| Ontology databases | BioPortal | bioportal.bioontology.org |
| Standards MIGS/MIMS | GSC, Genomic Standards Consortium | gensc.org |
Curation
For curation, imported data files were compiled, converted to tab separated value files (TAB) format and imported into base, a full-featured desktop database front end, provided by LibreOffice (https://no.libreoffice.org/).
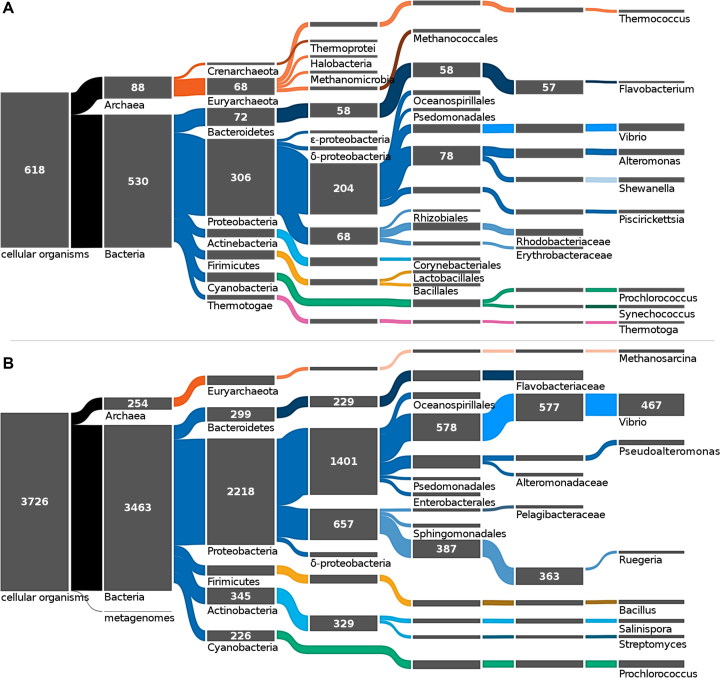
MarRef and MarDB contain in total 612 and 3726 records (Figure 2), respectively, with 106 metadata fields, out of which 30 fields are represented by CV and the remaining are free text or numeric fields. These 106 metadata fields include information about sampling environment, the organism and taxonomy, phenotype, pathogenicity, secondary metabolites, assembly and annotation.
Figure 2.
Most occurring marine taxa. (A) The reference database MarRef at its current state has 618 records of cellular organisms in the Archaea and Bacteria domains. Its complete and closed genomes are most prominent within the Proteobacteria phylum and the Alteromonadales order. (B) The partially curated database MarDB has 3726 records of sequenced genomes. Of its 287 unique genera (8 are shown) Vibrio is the most prominent with 467 records. These node-depleted Sankey diagrams were simplified to only display nodes exceeding 10 and 59 records for MarRef and MarDB respectively. An exception was made for the metagenome-derived genomes of MarDB.
The gene (protein) catalog database derived from marine metagenomic samples, MarCat, contains 1227 records, including samples from the Tara Ocean expedition (248 records) and Ocean Sampling Day (150 records). Each record contains 55 metadata fields.
The use of CV and ontologies can shortly be described by the following example. There are three environmental metadata fields used for describing the sampling site of a microorganism in MarRef and MarDB; environmental biome, feature and material which are controlled by a total of 95 terms. The environmental biome metadata field contains 11 controlled Environment Ontology (ENVO) terms covering environments such as Estuarine biome (ENVO:01000020), Marginal sea biome (ENVO:01000046), Marine benthic biome (ENVO:01000024), Marine mud (ENVO:00005795), Marine pelagic biome (ENVO:01000023), Marine water body (ENVO:00001999) and Ocean biome (ENVO:01000048). The environmental feature and material metadata fields are controlled by 59 and 25 terms, respectively. The ontologies used in the environmental biome, feature and material fields are all well-defined and described (http://www.environmentontology.org/), allowing consistency across the datasets.
The databases link out to other publicly available resources. For example, in MarRef sixteen of the metadata fields have active links to the literature databases such as PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) and PMC Europe (https://europepmc.org/), ontology databases such as ENVO (https://bioportal.bioontology.org/ontologies/ENVO) and Gazetteer (GAZ) (https://bioportal.bioontology.org/ontologies/GAZ), sequence databases such as the universal protein resource (UniProt) (http://www.uniprot.org/proteomes/) and ENA, taxonomy databases such as NCBI Taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy) and Silva (https://www.arb-silva.de/) and the Bacterial Diversity Metadatabase, BacDive (https://bacdive.dsmz.de/). Links to other external resources such as compound and secondary metabolites databases are provided if available. These links allow site visitors to easily access other web pages in order to obtain more information about each record.
For MarRef, all metadata fields have been manually curated to ensure consistency across the datasets, which allow the end user to easily search and filter records. While MarRef is thoroughly curated, MarDB and MarCat are only partly curated.
Records in the marine databases, MarRef, MarDB and MarCat follow the MlxS standard guidelines developed by the Genomic Standard Consortium, in addition to ontologies such as ENVO and GAZ.
Refinement and validation
OpenRefine (http://openrefine.org/) was used for refining the metadata fields by cleaning, trimming of leading and trailing whitespace, transforming data from one format into another and extending it with web services and external data. A validation tool was developed to convert the tab separated value files (TSV) to extensible markup language files (XML) and from TSV to XML to link the source TSV curation databases to the XML database. The validator defines a set of rules for the conversion–warnings and errors during conversion are reported.
SEQUENCE DATABASES
The MarRef, MarDB and MarCat sequence databases are based on the non-redundant genome and metagenome datasets obtained from ENA and NCBI and by manually inspection assigned as belonging to the ‘marine microbial biome’ according to our definition.
MarRef and MarDB
While MarRef is a database for completely sequenced marine prokaryotic genomes, MarDB includes all remaining sequenced marine prokaryotic genomes regardless the level of completeness. Both the MarRef and MarDB databases primarily built on gene, protein and genome sequences obtained from the Prokaryotic RefSeq Genomes database (18). All archaeal and bacterial genomes in RefSeq have been annotated using the NCBI’s Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) (25). However, ∼20% of all records in MarDB did not have any RefSeq entry with PGAAP annotations. Circumventing the lack of gene and protein information of these genomes, annotation was performed on pre-assembled sequences using Prokka, a command line software tool, for annotation of prokaryotic genomes (26).
MarCat
MarCat represents a catalog of uncultivable (and cultivable) full-length genes (proteins) derived from marine metagenomic samples based on the Marine projects in EBI metagenomics (https://www.ebi.ac.uk/metagenomics/). Metagenomic sequence reads were downloaded from ENA and annotated using META-pipe (https://arxiv.org/abs/1604.04103). In short, sequencing reads were merged, filtered and assembled using MEGAHIT (27), which has been shown to be one the best assemblers for metagenomic samples in the Critical Assessment of Metagenome Interpretation (CAMI) challenge (28). From the resulting contigs, full-length CDSs were predicted using MetaGeneAnnotator (29) and functionally assigned using a compilation of results from BLAST against UniRef (30), Priam (31) and InterProScan5 (32). Using META-pipe for gene prediction and functional assignment allowed us to generate a consistent catalog across the datasets in MarCat (See https://f1000research.com/articles/6–70/v1 for a more detailed description of functional assignment). As a start, we used the high-coverage and high-quality sequence outputs from the Tara Oceans and Ocean Sampling Day metagenomic projects (10,11). In addition, more than 30 projects of various sizes were included based on EBI’s marine projects. These were filtered in order to maintain the whole genome shotgun marine samples exclusively and also to avoid any project-interwoven freshwater samples. Some examples of these smaller projects include the Amazon continuum metagenomes (33) and western english channel diurnal study (34).
IMPLEMENTATION AND USER INTERFACE
The MAR databases have been incorporated into the Marine Metagenomics Portal (MMP) (https://mmp.sfb.uit.no/).
Contextual databases
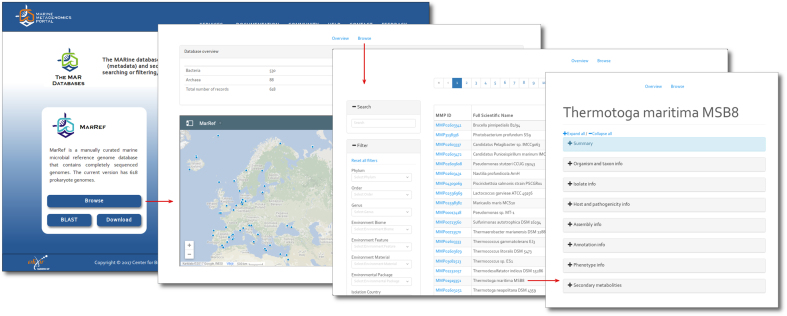
The contextual databases have been implemented using the hugo static website engine (https://gohugo.io/). The website engine reads the databases from XML files and allows the site visitor to access the information from four different layers. The first layer is the ‘Database selection’ page, where the user can select the different MAR databases for browsing, BLAST sequences or downloading (Figure 3). The second layer is the specific database ‘Overview’ page, which provides information about the content of the database and the geolocation of each genome/metagenome sample in the specific database. The geolocation has been embedded using google maps and each sample can be selected to display the organism/metagenome sample name and a short description of the organism/sample. The corresponding contextual information of the record can be reached by activating the MMP_ID link. The third layer is the ‘Browse’ (Figure 4) page which can be reached from the ‘Overview’ page and allows the site visitors to:
Browse the database records of interest.
Search across all metadata fields e.g. search for a specific organism, environmental ontologies, accession ID or any word.
Filter records to be visible in the table based on the most important record attribute, such as taxonomy (phylum, order and genus) and environmental ontologies (biome, feature and material).
Advanced filtering allows the site visitor to (a) add one or more filters; (b) refine current filters by adding new filters or removing already applied filters, (c) combine search and filtering and (d) remove all filters and launch a new search.
Figure 3.
Accessing the MAR databases and their records. From within the front page of the MMP all three metadatabases and sequence databases can be reached by following the ‘Browse’ or ‘BLAST’ buttons respectively. Browsing a metadatabase leads to the map-overview before reaching its index table. Single entries can be studied by selecting them in the map or in the table.
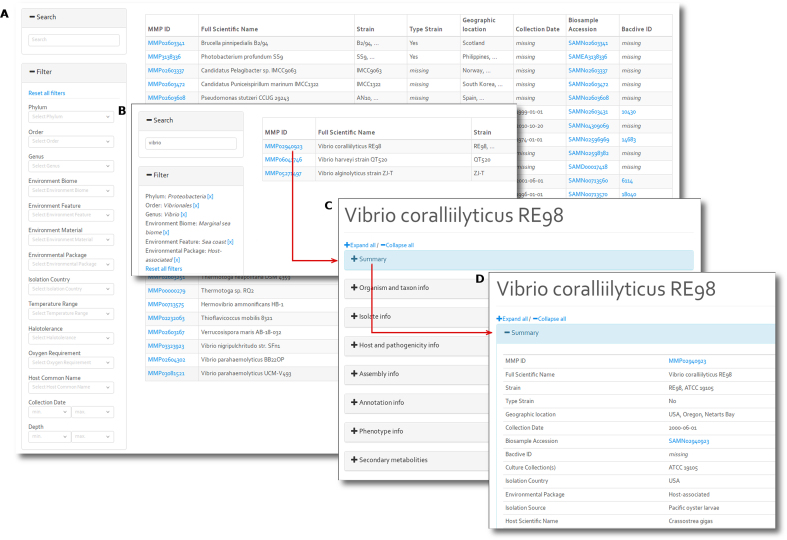
Figure 4.
The browsing interface and filtering functionality of MarRef and MarDB. (A) The default view as accessed from the corresponding database overview menu. The table content is instantaneously updated when filtering and responds to search words and 14 filtering fields. (B) Combining search words and filters enables search criteria to narrow the listed results in a highly flexible manner. (C) The metadata of each record is separated in eight expandable categories, (D) here illustrating parts of the summary. The index of MarCat (not shown) is less comprehensive, thus have fever filtering options.
The search/filtered results will be listed in a table. Summary of the metadata will be shown when activating the ‘Summary’ button. The fourth layer contains the information for each record. The contextual data for a record can be viewed using the ‘expand all’ button. For the marine genome databases, MarRef and MarDB, the 106 metadata fields in the record is divided into seven categories; organism and taxon info, isolate info, phenotype info, secondary metabolites, host and pathogenicity info, assembly info and annotation info, in addition to Summary. For MarCat, the metagenome databases, the 55 metadata fields have been divided into four categories: isolate info, sampling info, host and pathogenicity info and assembly info, in addition to Summary.
BLAST
The BLAST (35) sequence databases provide similarity search against all nucleotide and protein sequences of records included in MarRef, MarDB and MarCat. The BLAST functionality was established using SequenceServer Version 1.09 (https://doi.org/10.1101/033142) to provide the graphical user interface for the search results. The SequenceServer allows the visitor to type, paste or drag-and-drop a FASTA file to search either a single or several databases. The interface automatically recognizes the sequence type and chooses the appropriate BLAST method and databases. Advanced parameters (command line) can be used to refine the search. The output of BLAST consists of a list of hits with the corresponding E-value, and a set of the traditional pairwise alignments were the target sequence can be viewed and downloaded. From the pairwise alignment the visitor can also retrieve information of the organism/metagenome sample in the MAR databases by opening the mmp button. In MarRef and MarDB information about the targets sequences can be obtained by opening the NCBI button. For MarCat, the marine metagenomics gene catalogue, target information can be obtained from other databases such as UniProt, InterPro and Brenda. Output from the BLAST search can be downloaded in FASTA, XML files or tab-separated files (TSV) format.
Download
The download section accommodates the contextual databases, individual genome and metagenome related sequences, and BLAST databases. Contextual information for all entries/samples exists as TSV and XML files which are available for the current and prior release versions. In MarRef and MarDB sequences of individual genomes are grouped according to their names and contained in separate folders where assembly, nucleotide and protein data are accessible as FASTA files. A general feature format file is also provided for each genome. The full collection of contigs/scaffolds, nucleotide and protein sequences for the BLAST databases are accessible in the same directory tree and may also be downloaded freely. For samples in the MarCat database, all predicted 16 S sequences and assembled contigs in FASTA format can be downloaded. In addition, an output file from META-pipe containing all annotated contigs in the sample is also provided together with the individual predicted genes and protein sequences in FASTA format.
ONGOING DEVELOPMENTS
The ongoing activities can be classified into three broad categories: (i) acquisition of data, (ii) ontologies and CV and (iii) linked data and interoperability
Acquisition of sequence and contextual data
The collection of data from publicly available resources will continue. However, due to increasing amount of genomic and metagenomic sequence- and metadata, development of automatic and semi-automatic import tools that generate metadata for the curation database will be improved in order to build more efficient import pipelines. In this first version of the MarRef and MarDB databases, only prokaryote genomes have been included. In the future, we aim to include virus, eukaryote microbial genomes and transcriptome data. In addition, we aim to include metatranscriptomics data to enhance the quality of the MarCat.
Ontologies and controlled vocabularies
To enhance the curation efficiency and to provide a better reliability of the datasets, the number of metadata fields will be increased with ontologies and CV. This effort will not only streamline the manual curation, but also provide data robustness and easier aggregation and analysis. For MarCat we intend to include metadata fields for the provenance of analysis according to the recommendation by Hoopen et al. (24), which includes metagenomics analysis metadata such as filtering, assembly, taxonomy, gene prediction and functional assignment.
Linked data and interoperability
In order to expose and share the curated data, we are currently working together with EMBL-EBI to link the MAR database records to the BioSample and INSDC databases. To improve data interoperability, we intend to implement schema.org markup, so that MMP websites and services contain more structured information. This structured information will make it easier for the end user to discover, collate and analyze our data. We also aim to improve better systems for downloading single records or multiple records selected by searching or filtering of the datasets.
These improvements will be implemented in the next version of the databases scheduled for March 2018.
The functionality of the databases has been tested using different platforms and web browser, such as Safari, Firefox, Chrome and Edge, without any problems. We welcome user feedback by email to mmp@uit.no.
ACKNOWLEDGEMENTS
We thank all the experts who contributed to the MMP database annotations from their field of expertise, especially ELIXIR EXCELERATE WP6 partners. The MMP is part of the Norwegian node of the European ELIXIR project.
FUNDING
ELIXIR EXCELERATE funded by the European Commission within the Research Infrastructures programme of Horizon 2020 [676559]; Research Council of Norway [208481]; UiT The Arctic University of Norway. Funding for open access charge: UiT The Arctic University of Norway.
Conflict of interest statement. None declared.
REFERENCES
- 1. Arrigo K.R. Marine microorganisms and global nutrient cycles. Nature. 2005; 347:349–455. [DOI] [PubMed] [Google Scholar]
- 2. Creer S., Deiner K., Frey S., Porazinska D., Pierre Taberlet P., Thomas W.K., Potter C., Bik H.M.. The ecologist’s field guide to sequence-based identification of biodiversity. Methods Ecol. Evol. 2016; 7:1008–1018. [Google Scholar]
- 3. Zettler L.A., Artigas L.A., Baross J., Loka Bharathi P.A., Boetius A., Chandramohan D., Herndl G., Kogure K., Neal P., Pedrós‐Alió C. et al. McIntyre A. A global census of marine microbes. Life in the World's Oceans: Diversity, Distribution, and Abundance. 2010; Oxford: Wiley-Blackwell; 233–245. [Google Scholar]
- 4. Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R., Bult C.J., Tomb J.F., Dougherty B.A., Merrick J.M. et al. Whole-genome random sequencing and assembly of Haemophilus influenzae. Science. 1995; 269:496–512. [DOI] [PubMed] [Google Scholar]
- 5. Ishoey T., Woyke T., Stepanauskas R., Novotny M., Lasken R.S.. Genomic sequencing of single microbial cells from environmental samples. Curr. Opin. Microbiol. 2008; 11:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eloe-Fadrosh E.A., Paez-Espino D., Jarett J., Dunfield P.F., Hedlund B.P., Dekas A.E., Grasby S.E., Brady A.L., Dong H., Briggs B.R. et al. Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs. Nat. Commun. 2016; 7:10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hedlund B.P., Dodsworth J.A., Murugapiran S.K., Rinke C., Woyke T.. Impact of single-cell genomics and metagenomics on the emerging view of extremophile ‘microbial dark matter’. Extremophiles. 2014; 18:865–875. [DOI] [PubMed] [Google Scholar]
- 8. Gross L. Untapped bounty: sampling the seas to survey microbial biodiversity. PLoS Biol. 2007; 5:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laursen L. Spain’s ship comes. Nature. 2011; 475:16–17. [DOI] [PubMed] [Google Scholar]
- 10. Rusch D.B., Halpern A.L., Sutton G., Heidelberg K.B., Williamson S., Yooseph S., Wu D., Eisen J.A., Hoffman J.M., Remington K. et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007; 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sunagawa S., Karsenti E., Bowler C., Bork P.. Computational eco-systems biology in Tara Oceans: translating data into knowledge. Mol. Syst. Biol. 2015; 11:809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wommack K.E., Bhavsar J., Polson S.W., Chen J., Dumas M., Srinivasiah S., Furman M., Jamindar S., Nasko D.J.. VIROME: a standard operating procedure for analysis of viral metagenome sequences. Stand. Genomic Sci. 2012; 6:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mitchell A., Bucchini F., Cochrane G., Denise H., ten Hoopen P., Fraser M., Pesseat S., Potter S., Scheremetjew M., Sterk P. et al. EBI metagenomics in 2016 - an expanding and evolving resource for the analysis and archiving of metagenomic data. Nucleic Acids Res. 2016; 44:D595–D603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen I.A., Markowitz V.M., Chu K., Palaniappan K., Szeto E., Pillay M., Ratner A., Huang J., Andersen E., Huntemann M. et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017; 45:D507–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meyer F., Paarmann D., D'Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A. et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008; 9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glöckner F.O., Joint I.. Marine microbial genomics in Europe: current status and perspectives. Microb. Biotechnol. 2010; 3:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mineta K., Gojobori T.. Databases of the marine metagenomics. Gene. 2016; 576:724–728. [DOI] [PubMed] [Google Scholar]
- 18. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Verezemska O., Isbandi M., Thomas A.D., Ali R., Sharma K., Kyrpides N.C. et al. Genomes OnLine Database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 2017; 45:D446–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C., Conrad N., Dietrich E.M., Disz T., Gabbard J.L. et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017; 45:D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vallenet D., Calteau A., Cruveiller S., Gachet M., Lajus A., Josso A., Mercier J., Renaux A., Rollin J., Rouy Z. et al. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 2017; 45:D517–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kottmann R., Kostadinov I., Duhaime M.B., Buttigieg P.L., Yilmaz P., Hankeln W., Waldmann J., Glöckner F.O.. Megx.net: integrated database resource for marine ecological genomics. Nucleic Acids Res. 2010; 38:D391–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Costello M.J., Bouchet P., Boxshall G., Fauchald K., Gordon D., Bert W., Hoeksema B.W., Poore G.C.B., van Soest R.W.M., Stohr S. et al. Global coordination and standardisation in marine biodiversity through the World Register of Marine Species (WoRMS) and related databases. PLoS One. 2013; 8:e51629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoopen P.T., Finn R.D., Bongo L.A., Corre E., Fosso B., Meyer F., Mitchell A., Pelletier E., Pesole G., Santamaria M. et al. The metagenomic data life-cycle: standards and best practices. GigaScience. 2017; 6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Angiuoli S.V., Gussman A., Klimke W., Cochrane G., Field D., Garrity G., Kodira C.D., Kyrpides N., Madupu R., Markowitz V. et al. Toward an online repository of standard operating procedures (SOPs) for (meta)genomic annotation. OMICS. 2008; 12:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014; 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- 27. Li D., Liu C.M., Luo R., Sadakane K., Lam T.W.. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015; 31:1674–1676. [DOI] [PubMed] [Google Scholar]
- 28. Sczyrba A., Hofmann P., Belmann P., Koslicki D., Janssen S., Dröge J., Gregor I., Majda S., Fiedler J., Dahms E. et al. Critical assessment of metagemome interpretation—a benchmark of metagenomics software. Nat. Methods. 2017; doi:10.1038/nmeth.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noguchi H., Taniguchi T., Itoh T.. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008; 15:387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suzek B.E., Wang Y., Yuqi, Huang H., McGarvey P.B., Wu C.H. the UniProt Consortium . UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015; 31:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Claudel-Renard C., Chevalet C., Faraut T., Kahn D.. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2008; 31:6633–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones P., Binns D., Chang H-Y, Frase M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Satinsky B.M., Zielinski B.L., Doherty M., Smith C.B., Sharma S., Paul J.H., Crump B.C., Moran M.A.. The Amazon continuum dataset: quantitative metagenomic and metatranscriptomic inventories of the Amazon River plume, June 2010. Microbiome. 2014; 2:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gilbert J.A., Meyer F., Schriml L., Joint I.R., Mühling M., Field D.. Metagenomes and metatranscriptomes from the L4 long-term coastal monitoring station in the western english channel. Stand. Genomic Sci. 2010; 3:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L.. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008; 36:W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]