Abstract
The Library of Integrated Network-based Cellular Signatures (LINCS) program is a national consortium funded by the NIH to generate a diverse and extensive reference library of cell-based perturbation-response signatures, along with novel data analytics tools to improve our understanding of human diseases at the systems level. In contrast to other large-scale data generation efforts, LINCS Data and Signature Generation Centers (DSGCs) employ a wide range of assay technologies cataloging diverse cellular responses. Integration of, and unified access to LINCS data has therefore been particularly challenging. The Big Data to Knowledge (BD2K) LINCS Data Coordination and Integration Center (DCIC) has developed data standards specifications, data processing pipelines, and a suite of end-user software tools to integrate and annotate LINCS-generated data, to make LINCS signatures searchable and usable for different types of users. Here, we describe the LINCS Data Portal (LDP) (http://lincsportal.ccs.miami.edu/), a unified web interface to access datasets generated by the LINCS DSGCs, and its underlying database, LINCS Data Registry (LDR). LINCS data served on the LDP contains extensive metadata and curated annotations. We highlight the features of the LDP user interface that is designed to enable search, browsing, exploration, download and analysis of LINCS data and related curated content.
INTRODUCTION
Since the completion of the Human Genome Project (1), increasingly powerful technologies for large-scale multi-omics profiling and integrative data analytics have transformed the ability to gain insights and model the complex and dynamic networks of molecules and biological processes involved in human disease (2). Increasingly large and diverse datasets have empowered team-based research and big data approaches to model complex biological systems and drug action (3,4). Accordingly, systems biology and big data are also transforming drug discovery (5–7). The Library of Integrated Network-based Cellular Signatures (LINCS) project (http://lincsproject.org/), initiated by the NIH in 2011, aims to catalyze systems-biology approaches to model complex diseases and the development of novel therapeutics via an extensive catalog of perturbation response signatures in a wide array of cell model systems. The current LINCS Phase 2 includes six Data and Signature Generation Centers (DSGCs) and one Data Coordination and Integration Center (DCIC). The LINCS catalog of signatures can be thought of as a data cube, with three main dimensions: (i) the cell model system, including proliferating immortal cell lines, primary cells, and induced pluripotent stem cells (iPSCs); (ii) the perturbation, including small molecules, genetic perturbations, and microenvironment effectors and (iii) the cellular readout, including gene expression, phosphoproteins, epigenetic states, and various cellular phenotypes including morphology, signaling and cell fates, global proteomics, and biochemical small molecule-protein binding.
Another goal of LINCS is to develop information management and analysis software tools to make LINCS data accessible and useful to the research community. In contrast to other large-scale data generation efforts, the LINCS DSGCs employ a wide range of assay technologies of more than 20 high-throughput assays, posing a significant challenge to data processing and integration. To accomplish this goal, rigorous metadata specifications and data exchange standards were developed in the Pilot Phase and are maintained and expanded as new assays, cells, and perturbations are introduced in the Consortium (8). LINCS data standards are based on community best practices (9) to enable the linking and integration of LINCS to other resources.
All LINCS data have been organized via the LINCS Data Registry (LDR) after extensive data standardization and annotations with reference ontologies and identifiers so that all datasets have consistent metadata, enabling their integration and mapping to many external resources. The LINCS Data Portal (LDP) provides a unified interface to access LINCS datasets and metadata contained within the LDR. The LDP provides various options to explore, query, and download LINCS dataset packages and related objects, such as metadata for small molecules and cell lines utilized in the experiments.
In addition to managing the large quantity and diversity of data and extensive metadata records and reference annotations, the implementation of the LDP and design of the underlying registry were guided by the FAIR principles to make digital research objects Findable, Accessible, Interoperable, and Reusable (10) and by the Joint Declaration of Data Citation Principles (JDDCP) (Force11 Data Citation Synthesis Group (2014); https://www.force11.org/datacitation) to cite (attribute) LINCS datasets to their authors. Here we describe for the first time the LDR and the LDP as the two main resources to manage LINCS data and make LINCS resources accessible to the community.
MATERIALS AND METHODS
LINCS content overview
The LINCS Consortium produces data through the use of numerous assays and technologies, including multiple -omics categories (proteomics, transcriptomics, epigenomics), imaging, and the microenvironment microarray assay (MEMA), a novel platform for evaluating the effect of combining microenvironment perturbagens (e.g. growth factors, extracellular matrix proteins) on specific biological endpoints such as proliferation, differentiation, DNA damage, and senescence. High-throughput, large-scale assays include RNA-seq, L1000 (A Next Generation Connectivity Map: L1000 platform and the first 1,000,000 profiles: https://www.biorxiv.org/content/early/2017/05/10/136168), assay for transposase accessible chromatin sequencing (ATAC-seq), fluorescence microscopy, P100, global chromatin profiling (GCP), reverse phase protein array (RPPA), and sequential windowed acquisition of all theoretical fragment ion mass spectra (SWATH-MS). A full list of LINCS assay technologies and assays is provided in Supporting Table S1. In addition to various assays and corresponding data types, the LINCS data matrix expands over numerous cell types and perturbagens. Over 1000 cell types have been used in LINCS experiments, including established cell lines, embryonic stem (ES) cells, induced pluripotent stem cells (iPSCs), differentiated cells, and primary cells. Most are cancer cell lines covering a wide range of cancers such as melanoma, leukemia, lung, colon and breast carcinoma. Additionally, primary cells, ES cells, iPSCs, and differentiated cells are used in numerous disease modeling experiments, such as iPSCs derived from amyotrophic lateral sclerosis (ALS) and spinal muscular atrophy (SMA) patients and the corresponding in vitro differentiated motor neurons. The cells are treated with small molecules (which represent the largest group of perturbagens, with ∼42 000 small molecules tested), protein ligands, micro-environments, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (CAS9) mediated gene editing, or other perturbagen reagents or environmental factors. The small molecule collection covers a wide range of chemical perturbagens such as FDA-approved drugs, tool compounds (or chemical probes), and screening library compounds including those with clinical utility, known Mechanism of Action (MoA), and compounds previously tested in the NIH Molecular Libraries Program (11).
LINCS data for both experimental reagents and assays are standardized according to the previously established LINCS data standards (8), incorporating annotations from various ontologies, such as Cell Line Ontology (CLO) (12) for cells, Chemical Entities of Biological Interest (ChEBI) (13) for small molecules, and the BioAssay Ontology (BAO) (14,15) for assays. A full list of these ontologies, standards, and associated LINCS data/metadata elements is provided in Supporting Table S2. These data are used to construct the primary DCIC data product, the dataset packages, which contain datasets, their metadata, and any other related digital objects, e.g., property mapping and readme files. These comprehensive, data-analysis-ready packages are available for download as .tar groups from the LDP Landing Pages described below.
External data sources for value-added annotation and validation
LINCS data standardization, including reference ontologies, enable the mapping, validation, and incorporation of value-added information from various external repositories, including UniProt (16), PubChem Compounds (17), ChEMBL (18), and Cellosaurus (http://web.expasy.org/cellosaurus/). A comprehensive list of these external resources is provided in Supporting Table S3. To automate this integration, extract-transform-load (ETL) pipelines were built for the external data repositories including cleaning, aggregation, and data-type conversions before mapping to the LINCS research objects (e.g. small molecules, cell lines, protein targets, etc.). Examples include pipelines that garner MoA annotations, controlled vocabulary disease terms, aggregated bioactivities (IC50, Kd, etc.), and clinical indications for small molecules and protein targets (see Supporting Table S3). Other pipelines were built to validate consistency in biological annotations such as donor sex, tissue, disease associations, cell markers, culture conditions, and naming for cell lines and other biological entities.
Architecture and data processing
The LDR is the central data repository for the LINCS-DCIC and is the primary data source for the LDP. Data and metadata from DSGCs are transferred and processed by the DCIC via a collection of tools and protocols (e.g. Dataset Submission Tool, Metadata Submission Tool, Application Programing Interfaces (APIs), data downloads, or via a Center Website). The process includes data standardization and annotations from external sources (see MATERIAL & METHODS, External data sources for value-added annotation and validation) via a suite of well-defined automated pipelines (specific to the data types), which have several quality control (QC) inspection and validation points. All data objects and metadata are then registered into the LDR.
The LDR schema is implemented as a relational database combining a standard relational schema with key-value store. The key-value store captures all metadata fields and attributes. This design provides flexibility as the metadata standards evolve, with the ability to ensure data integrity using a relational model for the well-defined connections among entities, datasets, Centers, assays, and projects. The digital objects in the LDR include LINCS datasets, dataset groups, small molecules, cell lines, proteins, nucleic acid reagents, and peptide probes. An Entity Relationship (ER) diagram for the entire LDR is available in Supporting Figure S1.
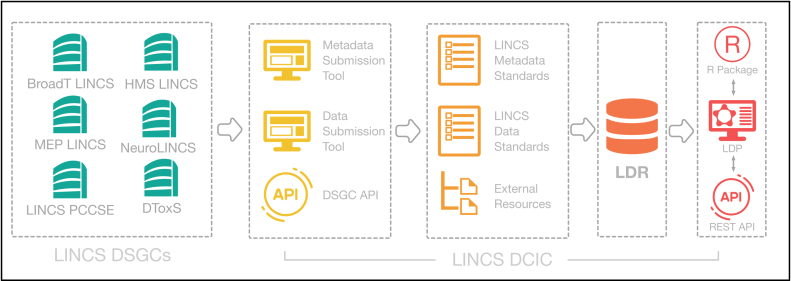
In order to achieve fluid and intuitive user experience in searching and navigating LINCS data, all data from the LDR are transformed and indexed using Apache Solr, a reliable and scalable search platform. Representational state transfer (REST) APIs have been developed using this Solr index to fetch data for display in the LDP and for public API accessibility (http://lincsportal.ccs.miami.edu/apis/), as illustrated in Figure 1. The LDP is a multi-tier, web-based application intended to function on a wide range of screens and devices. We have used open-source technologies for its three tiers: (i) PostgreSQL and Apache Solr to store data, (ii) Angular Javascript (JS) for presentation and (iii) Java-based servlets deployed on Tomcat as a service layer. For chemical structure search, we use the ChemAxon JChem PostgreSQL Cartridge (JChem Cartridge 2.6, 2017, ChemAxon, (http://www.chemaxon.com). Supporting Figure S2 illustrates a stack diagram of the LDP technologies and data flow.
Figure 1.
Data flow from LINCS DSGCs into the LDR and LDP. Data are submitted to the DCIC via different tools and then standardized, annotated, and loaded into the LDR. From the LDR, LINCS data are available via different mechanisms. Abbreviations: LINCS, Library of Integrated Network-based Cellular Signatures; DSGC, Data and Signature Generation Centers; DCIC, Data Coordination and Integration Center; HMS, Harvard Medical School; MEP LINCS, Microenvironment Perturbation LINCS Center; PCCSE, Proteomic Characterization Center for Signaling and Epigenetics; DToxS, Drug Toxicity Signature Generation Center; LDR, LINCS Data Registry; LDP, LINCS Data Portal; RESR, Representational State Transfer; API, Application Programming Interface.
RESULTS
Presentation and usability features
The LDP is designed to be broadly applicable and of use to both computational and non-computational scientists. The LDP includes several dedicated modules for exploring the complete collection of LINCS Datasets and the various associated objects, such as small molecules, cells, and other reagents and their metadata and annotations. These LDP modules have a summary page and a catalog browser with search functionality and facets for filtering content. For each category of research object, such as datasets or small molecules, the content is organized into dedicated (landing) pages (see below). The hierarchy of pages to organize and search/explore LINCS content is illustrated in Supporting Figure S3.
Augmenting the LDP is a REST API (http://lincsportal.ccs.miami.edu/apis/) that supports programmatic access to the search functionality and to all the data contained within the LDR. The API is designed to be self-describing with responses returned in JSON (JavaScript Object Notation) format. The API is of primary interest to computational scientists and developers building novel applications and research pipelines. We anticipate the most common interaction to be via the web interface; hence, here we focus on a description of features implemented in the web interface that enhance usability and exploration of the LINCS content.
Summary home page
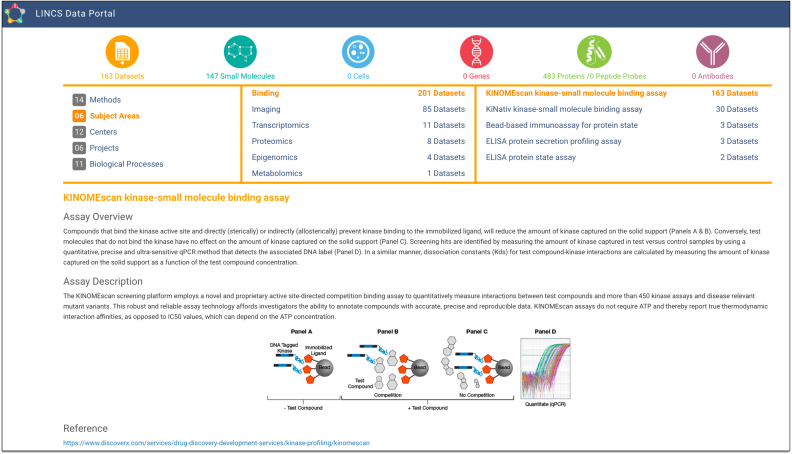
The entry point into the LDP is an interactive Summary Home Page (Figure 2). It allows users to get a quick overview of LINCS content based on categories, such as their respective methods (e.g. RNA-seq, ATAC-seq, L1000), biological processes (e.g. gene expression, cell proliferation), and general subject areas (e.g. transcriptomics, proteomics, epigenomics). For example, one can quickly find all binding assays, their corresponding datasets, and the number of tested small molecules (Figure 2). The LDP currently contains more than 350 datasets, almost 42 000 small molecules (including drugs and clinical compounds), and almost 1200 cells. The Summary Home Page links out specific modules including those for datasets, small molecules, or cells, and clicking on each symbol will direct the user to the list of respective objects corresponding to the selections in the Summary page. At the bottom, the home page shows brief descriptions and references for the selected LINCS assays, providing the user with a brief description of their selection.
Figure 2.
The LDP Summary Home Page is the entry point of LDP to interactively explore LINCS content based on several categories (http://lincsportal.ccs.miami.edu/dcic-portal/). Here, a specific selection of the KINOMEscan assay by Subject Area > Binding > KINOMEscan is shown, which contains 163 datasets and 147 small molecules.
Search functionality
The LDP provides various options to search, explore, and download the processed LINCS Dataset Packages, small molecules, cells, and other reagents along with their metadata and annotations. Free text search for all content is enhanced by autocomplete suggestions and grouped by various categories. The autocomplete feature is designed to be the primary entry point for exploring datasets and reagents. Ontologies, which are used to standardize and annotate LINCS content (see MATERIAL & METHODS, LINCS Content Overview), further expand the search. For example, while typing text, suggestions for drug names, biological targets, mechanisms of action, disease terms, etc., are made. The LDP also provides chemical structure search, enabling users to draw or paste/import .mol files to find small molecules (and associated datasets) by chemical substructure or topological similarity.
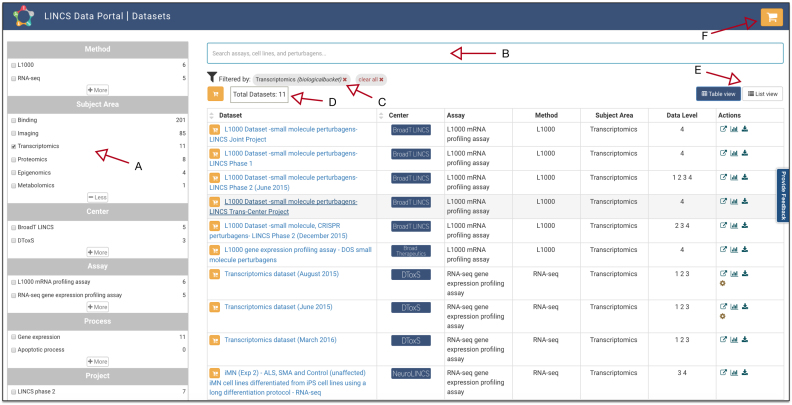
Facet filter controls provide simple and intuitive means of filtering search results for datasets and reagents. Facet-based filtering can be used to filter datasets by various categories, such as general study/subject area, assay method, assay name, biological process, center name, etc. For example, by selecting the ‘Transcriptomics’ area of study, the dataset list is instantly reduced to 11 from the complete list of 350 (Figure 3). The results can be further filtered using other facets or keywords. Both text search and facet filters are enabled by rich metadata annotations and the use of ontologies and controlled vocabularies for identifying the reagents and assays. Selecting a filter will automatically display the matching list of datasets, and multiple filters may be combined using a logical OR within a panel and logical AND in between panels. The count of datasets is dynamically updated and displayed next to each filter. The selected filters and search terms are displayed as tags below the search bar, and can quickly be individually removed or all cleared. Combining filters allows the construction of complex queries. Search results can be viewed in Table and List views, which can be toggled via buttons below the search bar. The tabular view, which is the default, provides a summary of each dataset including the name of the Center that generated the dataset, the assay name, assay method, available data levels, and various other summaries (as action links), such as count of reagents (small molecules, cells), link to the source, visualizations of dataset, and a download. Clicking on the dataset links to the Dataset Landing Page.
Figure 3.
Overview of the LDP catalog: (A) facets for filtering, (B) datasets count, (C) selected filters, (D) search bar for text search, (E) toggle between the views, (F) shopping cart for dataset bulk download.
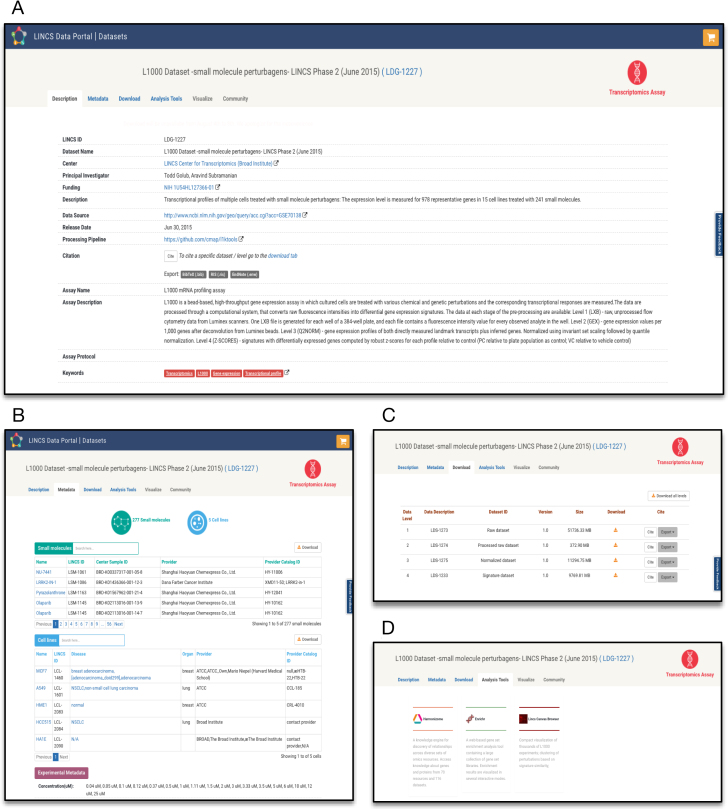
Dataset landing pages
Among the central features of LDP are the Dataset Landing Pages. Because LINCS signatures are generated via several stages (data levels) starting from the raw data, the Dataset Landing Pages refer to a dataset group that includes all data levels derived from an experiment. LINCS data levels typically range from level 1 to level 4, from raw instrument-produced data points to derived cellular signatures (Table 1). Each such LINCS dataset group is assigned a landing page by which all relevant information to the corresponding data are presented. The format of the landing pages is uniform across all LINCS dataset groups and currently consists of four sections (tabs): Description, Metadata, Download, and Analysis Tools (Figure 4). The Description tab provides important details describing how and by whom the dataset was generated and processed including a description and protocol of the assay, relevant publications, and standardized keywords. The Metadata tab contains searchable and downloadable information regarding the research objects associated with the dataset, such as small molecules, proteins and cells. Links to relevant external resources are also available in this section.
Table 1. LINCS Data level definitions, descriptions and examples.
| Data Level | Data Type | Description | Examples |
|---|---|---|---|
| 1 | Raw | Non-normalized, low-level, single-sample data obtained from assay instrumentation | RNA-Seq FASTQ, L1000 LXB |
| 2 | Processed | Raw data converted to a basic usable format through data processing, including presence or absence of specific molecular abnormalities | KiNativ binding profile, KINOMEScan relative inhibition |
| 3 | Normalized | Normalized, per sample data | L1000 Q2NORM, RNA-Seq raw counts |
| 4 | Signature | Quantified association across samples based on two or more characteristics | RNA-Seq Differential expression profile, L1000 GCTX file |
Figure 4.
LINCS Dataset Landing Page: (A) Dataset Description tab, (B) Associated Metadata tab, (C) Download tab, (D) Analysis tools tab. The screenshots correspond to dataset LDS-1233 (http://lincsportal.ccs.miami.edu/datasets/#/view/LDS-1233).
To enable the integration of diverse LINCS data and to provide a common structure of datasets produced by different Centers using diverse technologies and data processing pipelines, the data are organized into standardized dataset packages. A dataset, in this context, refers to one specific data level (in contrast, the group includes the corresponding datasets for all levels). Each dataset package contains: (i) a readme file with all the relevant annotations regarding the dataset, (ii) the original data files as produced by the DSGC, (iii) the standardized metadata of all the reagents used in the dataset, with one file per category (e.g. small molecule, cell, protein) and (iv) mapping files that associate the metadata and data files. These files are created by secondary data processing, standardization, annotation, and curation (see MATERIAL & METHODS, Architecture and data processing). Because metadata are all standardized with unique identifiers, different datasets can easily be mapped and integrated. The LINCS data packages are available to download via the Download tab. Citations for the different dataset levels are also made available in the Download tab. Finally, the Analysis Tools tab includes a list of the computational tools that allow the exploration and analysis of the dataset as well as links to pre-generated reports containing the results of analyses that have been performed on the dataset.
To cite LINCS data, we created collections of LINCS digital research objects (DROs) in the Minimal Information Required in the Annotation of Models (MIRIAM) Registry that generate unique, perennial, and location-independent identifiers (http://identifiers.org/) (19). Such collections include data-level specific dataset packages, dataset groups (all data levels), small molecules, and cells. The identifiers.org service, which is built upon the information stored in MIRIAM, provides directly resolvable identifiers in the form of Uniform Resource Locators (URLs). This system provides a globally unique identification scheme to which any external resource can point and a resolving system that gives the owner/creator of the resource collection flexibility to update the resolving URL without changing the global identifiers. These dataset and dataset group identifiers are the central component of the LINCS dataset citation record, which further includes the authors, title, year, repository, resource type, and version. These citation records have been incorporated into the LINCS Data Portal and can be downloaded in several formats, making it easy to cite a specific LINCS dataset or dataset group. All LINCS datasets are indexed in bioCADDIE DataMed (https://datamed.org/) and OmicsDI (http://www.omicsdi.org/), which link back to the dataset landing pages via their global IDs.
Dataset download and shopping cart
LDP supports individual dataset download from the Dataset Landing Page and also bulk download using the shopping cart that allows the addition of selected datasets directly from the catalog page. Bulk download is particularly useful to access related datasets, such as those generated by the same assay. The shopping cart downloads the highest dataset level available, which is what typical end users want, and also contains a Python script that facilitates merging the various files into a single file based on the assays and data format.
LDP small molecule module
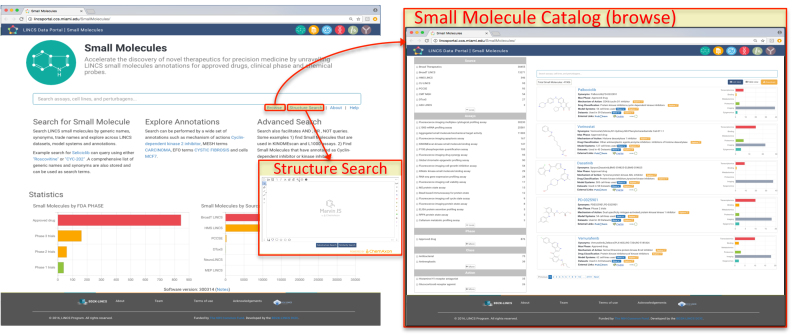
The Small Molecule Landing Page (http://lincsportal.ccs.miami.edu/SmallMolecules/) provides an overview page with statistics displayed by FDA clinical phase and source, a prominent search window, and links to the catalog (browse) and chemical structure search (Figure 5). Before registration into the LDP, LINCS small molecules were standardized following business rules at the DCIC so that different variations of the same compound (such as salt forms, tautomers, and ionization states), but not different chemical structures (e.g. stereoisomers or geometric E/Z isomers), receive the same unique ID to enable data integration. Standardized small molecules are currently cross-referenced to PubChem Compounds (17) (via CID), ChEMBL (18), ChEBI (13), and UniChem (https://www.ebi.ac.uk/unichem/), which provides cross-reference to 37 additional public databases. In addition, small molecules were extensively annotated and curated with characteristics including FDA approval status, clinical trial phases, mechanisms of action, biochemical targets, clinical disease indications, and many computed drug-relevant properties. The small molecule pages are integrated with the datasets so that all datasets in which a drug was tested are immediately available and cross-linked. The database also keeps track of the original specific samples tested at the DSGCs. Small molecules can be searched by (drug) name, MOA, biological target, clinical disease indication, and by free text and chemical structure. The small molecule catalog allows browsing and filtering via various facets, including MOA, clinical phase, and pharmacological class. Similar to datasets, each small molecule has a landing page with a unique URL (which resolves globally via identifiers.org). Each small molecule landing page consists of several sections including Description with basic information (name, synonyms, ID, cross-references, various descriptors), associated LINCS and external Datasets (with an overview by areas of study and cell types), and Curated Data including biological targets and diseases. Via the Description tab, a user can also search for structurally similar compounds.
Figure 5.
Left: Small Molecule Landing Page (http://lincsportal.ccs.miami.edu/SmallMolecules/) with statistics displayed by FDA clinical phase and source, a prominent search window, and links to the catalog (browse) and chemical structure search. Right: Small Molecule Catalog with chemical structures, names, and various annotations and links, statistics to the right, filter facets to the left, search window at the top.
Other modes of data access
APIs
To accommodate users who seek information about a single dataset, as well as computational biologists who can programmatically operate on the data, the LDP includes advanced search functionality, and serves the data in JSON format through an API (http://lincsportal.ccs.miami.edu/apis/).
R Package
In addition to the REST API, R users can also programmatically access the LDP through the R package LINCSDataPortal (https://github.com/schurerlab/LINCSDataPortal/). This package provides easy access to all the data and metadata that are stored in the LDP. More specifically, LINCS data packages can be retrieved using a variety of search terms for entities of interest (e.g. small molecule, protein, gene, cell line) and downloaded for any of the available data levels. Moreover, all metadata associated with any reagent used in LINCS experiments can also be retrieved.
DISCUSSION
The LDP organizes LINCS content by datasets and metadata categories via different modules that feature catalog and landing pages (see Supporting Figure S3). The LDP provides extensive search functionality, flexible browsing and filtering, and data download. Citation records for LINCS datasets can easily be created, leveraging unique global identifiers. In building the LDP and the underlying infrastructure and processes, particular emphasis has been placed on data standards including the use of ontologies, extensive curated annotations of LINCS content, globally valid identifiers, and easily accessible APIs. These efforts have been guided by the FAIR principles to make LINCS data Findable, Accessible, Interoperable, and Reusable (10). For example, LINCS data are already indexed in bioCADDIE DataMed (https://datamed.org/) and OmicsDI (http://www.omicsdi.org).
Via its current functionality, the LDP enables a wide range of use cases. For example, if the user wishes to discover all LINCS registered compounds that are indicated to treat glioblastoma (GBM), they can query the Small Molecule Application for the disease by the Medical Subject Heading (MeSH) term or Experimental Factor Ontology (EFO) annotations; this search returns 26 compounds. These compounds can be filtered by protein kinase inhibitors (via the pharmacological class facet), reducing the list to 11 compounds. Search results can be additionally filtered by approved drug (clinical Phase facet), leaving 9 drugs. The corresponding datasets for any of these compounds, including the complete kinase profile (KINOMEscan) or the transcriptional profile in various cells, can be downloaded. To continue this exploration, as epidermal growth factor receptor (EGFR) could be a relevant target in GBM, one can further filter the list by MoA target through the facet or a text search that will suggest EGFR as a target. That leaves one compound, afatinib. The available datasets and annotations are easily accessible through the compound's landing page. Any identified dataset can easily be cited via the Cite button on the Dataset Landing Page, and the citation can be downloaded in several formats. In the described manner, LDP functionality can be utilized in efforts for drug repositioning. As another approved drug studied for GBM, but not a kinase inhibitor, doxorubicin (LSM-4062) can easily be identified using a similar workflow. If a user is interested in related heterocycles with trapped quinones, one can search by chemical structure similarity directly in the doxorubicin page. The 13 compounds that are retrieved at 95% similarity include three additional approved drugs (epirubicin, daunorubicin, idarubicin), which are all also DNA topoisomerase inhibitors. These compounds could be purchased from commercial sources and screened for this activity, if desired. For better understanding, the described workflows are illustrated in Supplementary Figures S4 to S10.
The LDP is the primary access point to all data produced in the LINCS project, including the Pilot Phase and the current Phase 2. As described above, the LDP makes it easy for end users to browse, query, and filter LINCS data based on numerous categories and curated annotations, which were rigorously standardized to enable an integrated view across all LINCS content. The LDP also enables integration with external resources via added links, curated mappings, and external IDs. Content from many resources has been incorporated into the LDP. In addition to the Web-based user interface, LINCS data can easily be accessed via APIs and the R package. Global, perennial identifiers for LINCS datasets, as well as the most important related research objects and their mapping and description by community standard ontologies, are also critical components to ensure the persistence of LINCS data in the future. As the LINCS project progresses, many new released datasets will appear in the LDP, and we will add new features, including user-specific queries, lists, and settings, and further improve the annotations of LINCS content and integration with external data sources. The LDP makes LINCS data widely accessible thus contributing to the success of the LINCS program as a valuable scientific resource.
AVAILABILITY
The LINCS Data Portal (LDP) is freely available without restrictions at http://lincsportal.ccs.miami.edu/dcic-portal/.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the LINCS Consortium for feedback regarding LDP end user functionality and for reviewing and approving the representation of the hosted datasets and metadata in the LDP. We acknowledge resources from the Center for Computational Science (University of Miami, FL) for maintaining and administering the production environment and server for the LINCS Data Portal and related infrastructure. We thank ChemAxon Inc. for providing free academic research licenses, which were used in curating and annotating chemical structures, and a license to use the JChem PostgreSQL Cartridge that enables LDP chemical structure search.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Heart, Lung, and Blood Institute through funds provided by the National Institutes of Health Translational Research [U54HL127624]; Library of Integrated Network-based Cellular Signatures (LINCS) Program (http://www.lincsproject.org/); National Institutes of Health Translational Research Big Data to Knowledge (BD2K) initiative (http://www.bd2k.nih.gov/). Funding for open access charge: NIH [U54HL127624].
Conflict of interest statement. None declared.
REFERENCES
- 1. Collins F.S., McKusick V.A.. Implications of the human genome project for medical science. JAMA. 2001; 285:540–544. [DOI] [PubMed] [Google Scholar]
- 2. Sun Y.V., Hu Y.-J.. Integrative Analysis of Multi-omics Data for Discovery and Functional Studies of Complex Human Diseases. Adv. Genet. 2016; 93:147–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ma’ayan A., Rouillard A.D., Clark N.R., Wang Z., Duan Q., Kou Y.. Lean Big Data integration in systems biology and systems pharmacology. Trends Pharmacol. Sci. 2014; 35:450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xie L., Draizen E.J., Bourne P.E.. Harnessing Big Data for Systems Pharmacology. Annu. Rev. Pharmacol. Toxicol. 2017; 57:245–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Butcher E.C., Berg E.L., Kunkel E.J.. Systems biology in drug discovery. Nat Biotech. 2004; 22:1253–1259. [DOI] [PubMed] [Google Scholar]
- 6. Kiyosawa N., Manabe S.. Data-intensive drug development in the information age: applications of Systems Biology/Pharmacology/Toxicology. J. Toxicol. Sci. 2016; 41:SP15–SP25. [DOI] [PubMed] [Google Scholar]
- 7. Poornima P., Kumar J.D., Zhao Q., Blunder M., Efferth T.. Network pharmacology of cancer: From understanding of complex interactomes to the design of multi-target specific therapeutics from nature. Pharmacol. Res. 2016; 111:290–302. [DOI] [PubMed] [Google Scholar]
- 8. Vempati U.D., Chung C., Mader C., Koleti A., Datar N., Vidovic D., Wrobel D., Erickson S., Muhlich J.L., Berriz G. et al. . Metadata standard and data exchange specifications to describe, model, and integrate complex and diverse high-throughput screening data from the Library of Integrated Network-based Cellular Signatures (LINCS). J. Biomol. Screen. 2014; 19:803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McQuilton P., Gonzalez-Beltran A., Rocca-Serra P., Thurston M., Lister A., Maguire E., Sansone S.-A.. BioSharing: curated and crowd-sourced metadata standards, databases and data policies in the life sciences. Database. 2016; baw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E. et al. . The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Austin C.P., Brady L.S., Insel T.R., Collins F.S.. NIH molecular libraries initiative. Science. 2004; 306:1138–1139. [DOI] [PubMed] [Google Scholar]
- 12. Sarntivijai S., Lin Y., Xiang Z., Meehan T.F., Diehl A.D., Vempati U.D., Schürer S.C., Pang C., Malone J., Parkinson H. et al. . CLO: The cell line ontology. J. Biomed. Semantics. 2014; 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Degtyarenko K., de Matos P., Ennis M., Hastings J., Zbinden M., McNaught A., Alcántara R., Darsow M., Guedj M., Ashburner M.. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008; 36:D344–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abeyruwan S., Vempati U.D., Küçük-McGinty H., Visser U., Koleti A., Mir A., Sakurai K., Chung C., Bittker J.A., Clemons P.A. et al. . Evolving BioAssay Ontology (BAO): modularization, integration and applications. J. Biomed. Semantics. 2014; 5:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vempati U.D., Przydzial M.J., Chung C., Abeyruwan S., Mir A., Sakurai K., Visser U., Lemmon V.P., Schürer S.C.. Formalization, annotation and analysis of diverse drug and probe Screening Assay Datasets Using the BioAssay Ontology (BAO). PLoS One. 2012; 7:e49198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R. et al. . The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006; 34:D187–D191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. et al. . PubChem Substance and Compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B. et al. . ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012; 40:D1100–D1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Juty N., Le Novère N., Laibe C.. Identifiers.org and MIRIAM Registry: community resources to provide persistent identification. Nucleic Acids Res. 2012; 40:D580–D586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.