Abstract
SABIO-RK (http://sabiork.h-its.org/) is a manually curated database containing data about biochemical reactions and their reaction kinetics. The data are primarily extracted from scientific literature and stored in a relational database. The content comprises both naturally occurring and alternatively measured biochemical reactions and is not restricted to any organism class. The data are made available to the public by a web-based search interface and by web services for programmatic access. In this update we describe major improvements and extensions of SABIO-RK since our last publication in the database issue of Nucleic Acid Research (2012). (i) The website has been completely revised and (ii) allows now also free text search for kinetics data. (iii) Additional interlinkages with other databases in our field have been established; this enables users to gain directly comprehensive knowledge about the properties of enzymes and kinetics beyond SABIO-RK. (iv) Vice versa, direct access to SABIO-RK data has been implemented in several systems biology tools and workflows. (v) On request of our experimental users, the data can be exported now additionally in spreadsheet formats. (vi) The newly established SABIO-RK Curation Service allows to respond to specific data requirements.
INTRODUCTION
In 2006, SABIO-RK database (1) has been established to support modellers of biochemical reactions and complex networks. SABIO-RK represents a repository for structured, curated and annotated data about reactions and their kinetics. The data are manually extracted from the scientific literature and stored in a relational database. As compared with automatic data extraction by text mining tools, the manual extraction process guarantees a very high degree of accurateness and completeness. Especially, the extraction of the complex information of reactions and kinetics of most of the available publications are not enough structured and well written. Furthermore, relevant information is distributed over the entire article and unique identifiers or controlled vocabularies are missing (2,3). Based on the time consuming process of manual data extraction and manual curation, SABIO-RK emphasizes on quality rather than on quantity. SABIO-RK is not only a database for modellers but also for experimentalists in the laboratory who are looking for example for more details about the enzymatic activity of a protein or about alternative reactions of an enzyme. For many years SABIO-RK was focussing on kinetics of metabolic reactions but with an increased user interest in kinetics data for signalling events, SABIO-RK also stores reactions and binding events of signal transduction pathways.
The bidirectional cross-references between SABIO-RK and protein specific databases like UniProtKB (4), pathway databases like KEGG (5) or chemical compound databases like ChEBI (6) assist users to find more specific kinetic information in SABIO-RK and vice versa.
A comparable database providing kinetic parameters is BRENDA (7). In contrast to SABIO-RK, the information in BRENDA is centred on enzymes and their kinetic constants, whereas SABIO-RK focuses on reactions and additionally, beside constants, offers the associated kinetic rate laws, formulas and experimental conditions. Other databases containing kinetic data are focussing e.g. on proteins (UniProtKB), plant metabolism (MetaCrop (8)) or protein interactions (KDBI (9)).
NEW DATABASE CONTENT
The most SABIO-RK content sources are articles published between the late 1960s and today, which comprise currently more than 300 different journals. The selection of papers has changed over time. In the first years most of the publications were non-specifically selected by reaction kinetics related keyword search in the PubMed database (10), nowadays the focus of the selection is dependent on collaboration projects and user requests. The fact, that more than one third of the database content refers to mammals (mainly human and rat) and around 15% are liver data, is the result of such a former collaboration project. And that 25% of the data in SABIO-RK are related to the central Glycolysis/Gluconeogenesis pathway is due to user requests and several smaller projects. An increased user interest in plant metabolism is reflected in about 10% reaction kinetics data for green plants (embryophyta). All in all, the database content increased since the last NAR publication 2012 ∼40%. As of September 2017 SABIO-RK provides data extracted from more than 5.600 publications, stored in ∼57.000 different database entries. The kinetics data are related to 934 different organisms, of which about two-thirds belong to eukaryotes and one-third to bacteria, archaea and viruses. At present the top ten organisms in SABIO-RK are Homo sapiens, Rattus norvegicus, Escherichia coli, Saccharomyces cerevisiae, Mus musculus, Bos taurus, Bacillus subtilis, Arabidopsis thaliana, Sus scrofa and Oryctolagus cuniculus. A more detailed statistic about the database content is depicted on Figure 1.
Figure 1.
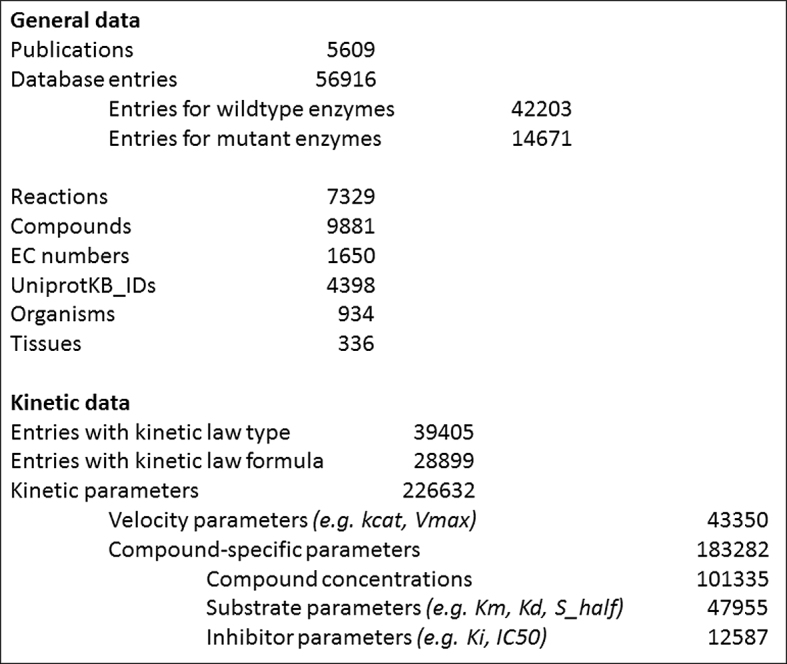
SABIO-RK statistics (status: September 2017).
SABIO-RK mostly contains metabolic reactions and only a small fraction for signalling and transport reactions. Currently there are kinetic data for ∼80 different signalling and 150 different transport reactions stored in the database. Transport reactions include reactions with and without chemical conversion of substrates. Reactions usually are assigned to pathways, which are based on the classifications from KEGG. But given that often alternative or non-biochemical compounds are used in experiments, there are many alternative reactions in SABIO-RK which are not linked to a biochemical pathway.
A database entry in SABIO-RK comprises kinetics data for one single reaction in one organism under specific experimental conditions. If the publication provides information for more than one biochemical reaction, organism or enzyme, these data will be stored not as one single, but in several distinct database entries. About 25% of the database entries contain data for specific mutant enzyme variants which allow the comparison of kinetics data from mutant with wildtype proteins.
More than 90% of SABIO-RK data have been manually extracted from publications. Biological experts read the paper and insert relevant information in a web-based curation interface where the data are semi-automatically checked for correctness and consistency. Annotations and unique identifiers are added for interoperability and interlinkage with ontologies, controlled vocabularies and external databases. Additionally, kinetics data from lab experiments or models can be directly uploaded into the curation interface via SBML format and further processed by the curators.
Data in the database entry and in the details pages are highly interlinked to external databases, ontologies and controlled vocabularies. Details pages for the reaction, organism, enzyme, pathway and compound are additionally shown in extra pop-up windows after clicking on the appropriate term. Links are implemented for reactions to KEGG, for proteins to UniProtKB, for organisms to NCBI taxonomy (10), for tissues to Brenda Tissue Ontology (BTO) (11), for publications to PubMed, for compounds to ChEBI, KEGG and PubChem (10), for cell locations and signalling events to Gene Ontology (GO) (12), for kinetic laws and parameters to Systems Biology Ontology (SBO) (13), and for enzymes to ExPASy (14), KEGG, BRENDA, IntEnz (15), IUBMB (http://www.chem.qmul.ac.uk/iubmb/enzyme/), Reactome (16) and MetaCrop.
NEW DATA ACCESS
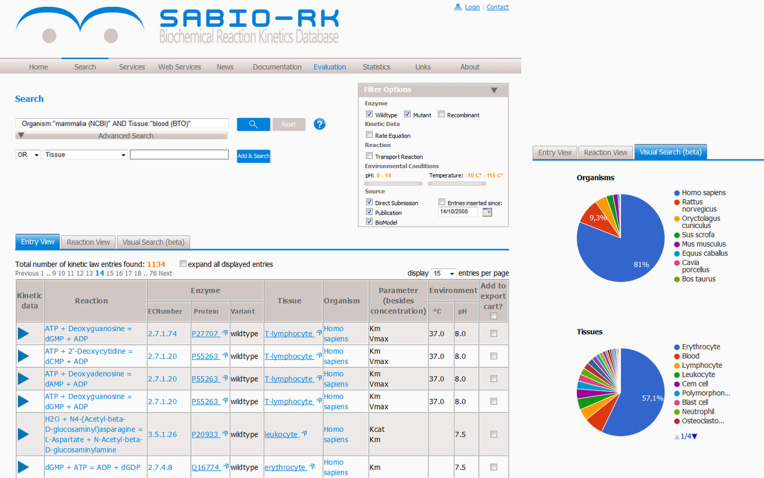
Data in SABIO-RK can be retrieved both via the web-based search interface and REST-ful web services. The most obvious change affects the website, which has been adapted to a more modern design, but also contains new features, like free text search. A free text search for ‘liver’ will return all entries containing this search term independent of the data field (e.g. tissue, publication title or comment). The advanced search feature allows the definition of complex queries by selecting different attributes like enzyme name, tissue, PubMedID, etc. from a selection list. This selection list includes not only names but also SABIO-RK internal as well as external identifiers (from KEGG, ChEBI, UniProtKB, GO, SBO etc.) and the possibility to search for signalling events (e.g. protein autophosphorylation) or signalling modifications (e.g. acetylation). The autocomplete function instantaneously makes suggestions and predicts how many results (database entries) are in the database for the given query. Figure 2 shows an example of the earlier introduced ontology-screened search for organisms using NCBI taxonomy and tissues using BTO and the results for classified groups of organisms and tissues in the new website design.
Figure 2.
Screenshots of SABIO-RK newly designed web-based search interface with query results for kinetics data in mammalian blood using ontology-based search (using NCBI taxonomy for organism and BTO for tissue).
Additional options can now be specified by defining filters in the filter options box. Filters can be set e.g. for enzymes/proteins by selecting data for wildtype or mutant proteins. Selecting the rate equation filter will display only data entries with a kinetic rate equation and accordingly, transport reactions are displayed when transport reaction filter is selected. The environmental conditions pH and temperature can be specified by moving the slider buttons to select a range. Since SABIO-RK contains data from different kinds of sources (publication, direct submission from laboratory or model upload via SBML) filter could be defined to search for specific data sources.
Search results are displayed in three different views: Entry view, Reaction view, and Visual search. By default the Entry view is shown, which lists the resulting database entries in a summarized way. Detailed information for each database entry can be viewed by clicking on the blue triangle. The Reaction view groups the database entries based on their reactions. To get a quick impression about the connection of a certain reaction with enzymes, organisms, and tissues a corresponding visualization is provided in this view. Columns in both, Entry and Reaction view can be sorted by clicking on the column header. Finally, the Visual search depicts a visualization of the search result together with the opportunity to confine the query by clicking on parts of the diagrams for organisms, tissues, kinetic parameters or kinetic rate laws. A partial screenshot of the Visual search containing the diagrams for organisms and tissues is shown in Figure 2.
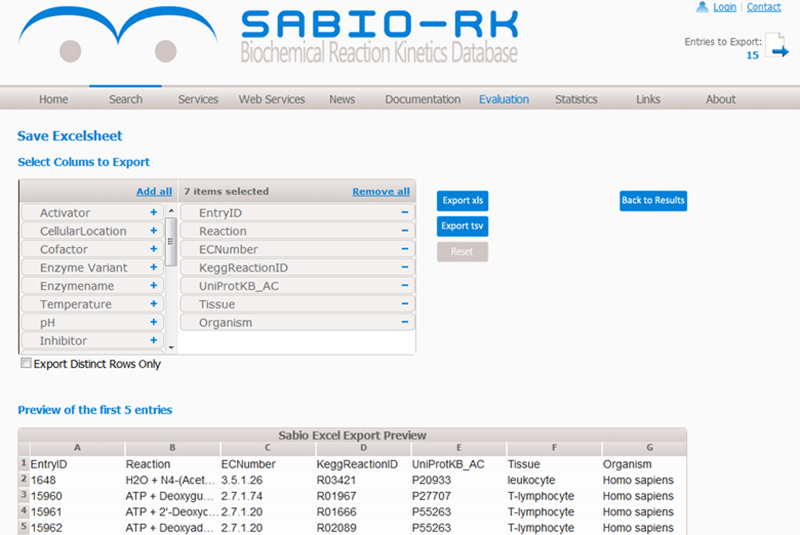
Search results of a SABIO-RK query can be selected for export by collecting database entries in an export cart. Data can be exported in standard exchange formats including SBML (17), BioPAX (18), SBPAX (19), MatLab (http://www.mathworks.com) and in spreadsheet format where the exported table columns can be defined by (de)selecting attributes from the list (see Figure 3).
Figure 3.
Screenshot of SABIO-RK web-based search interface representing spreadsheet export functionality.
Beside the web interface, SABIO-RK web services can be used to access the database automatically which is also used for retrieval of kinetics data by third-party software tools and data workflows. These tools include CellDesigner (20), VirtualCell (21), Sycamore (22), SBMLsqueezer (23), cy3sabiork (http://apps.cytoscape.org/apps/cy3sabiork), Path2Models (24), LigDig (25), FAIRDOMHub (26). Currently SABIO-RK is accessed mostly (ca. 90%) via web services, which underlines the importance of its integration in modelling and visualization tools.
Standard export formats for the web services are SBML, BioPAX/SBPAX and XML. Beside that a Python script is offered to use the web services for data export in table format.
SABIO-RK is cross-referenced by several other biological databases and online platforms which allows the users of these external resources to gain further knowledge about enzymatic activities of enzymes (links from UniProtKB, BRENDA, NextProt (27), ChloroKB (28), MetaCrop) and detailed information about kinetics of biochemical reactions (links from KEGG Reaction, MetaNetX (29), BKMS-react (30)) as well as the participation and meaning of compounds (links from ChEBI, MetaNetX) on it. Currently about 20% of SABIO-RK users are entering the database search interface through cross-references from external databases. External links are implemented using the same structure of query definition as in the search interface (e.g. http://sabiork.h-its.org/newSearch?q=ecnumber:2.7.1.40) to implement detailed or complex queries. The results of the query links can be further refined in the search interface.
NEW SERVICES
To adapt the SABIO-RK database even more to user requirements in regard to the database content, we support specific curation requests. In case that no results are returned for a specific query, the following note is displayed: ‘Sorry, we found no results for your query… — but you may send a request to add the corresponding data’, to encourage users to send their specific questions via the SABIO-RK contact form. For example SABIO-RK curators will help to find relevant data in the literature and insert the kinetics data extracted from the publications in the database. This service can include searches for kinetics data for specific organisms, pathways or enzymes. Here SABIO-RK is flexible enough and not restricted to any organism class or biochemical reaction type. This service is free of charge and a list of public curation request is displayed on a separate website for services (http://sabiork.h-its.org/publicCuration/list). Other types of requests, feedback or bug reports can always be given by using the contact form. Additionally, to foster an interactive exchange amongst users and between users and the SABIO-RK team, an internet forum via Google Groups has been established.
Users can also request for data upload of their own experimental data and models ideally given in SBML format. These data then run through the curation process, are annotated and linked to controlled vocabularies, ontologies and external databases, to allow the comparison of these private results with published data. Unpublished data or models can be protected from public access in a user password restricted area.
ACKNOWLEDGEMENTS
Over the years many people worked on SABIO-RK software and database development or contributed to the database content. Special thanks go to Meik Bittkowski, Lei Shi, Lenneke Jong, Elina Wetsch, Enkhjargal Algaa, Heidrun Sauer-Danzwith, Olga Krebs and Martin Golebiewski. The authors also would like to thank the database users and collaboration partners for permanent feedback and discussions about requirements and database improvements.
FUNDING
Klaus Tschira Foundation (http://www.klaus-tschira-stiftung.de/); German Federal Ministry of Education and Research (http://www.bmbf.de/) within de.NBI [031A540]. Funding for open access charge: HITS gGmbH (http://www.h-its.org).
Conflict of interest statement. None declared.
REFERENCES
- 1. Wittig U., Kania R., Golebiewski M., Rey M., Shi L., Jong L., Algaa E., Weidemann A., Sauer-Danzwith H., Mir S. et al. SABIO-RK – database for biochemical reaction kinetics. Nucleic Acids Res. 2012; 40:D790–D796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wittig U., Rey M., Kania R., Bittkowski M., Shi L., Golebiewski M., Weidemann A., Müller W., Rojas I.. Challenges for an enzymatic reaction kinetics database. FEBS J. 2014; 281:572–582. [DOI] [PubMed] [Google Scholar]
- 3. Wittig U., Kania R., Bittkowski M., Wetsch E., Shi L., Jong L., Golebiewski M., Rey M., Weidemann A., Rojas I. et al. Data extraction for the reaction kinetics database SABIO-RK. Perspect. Sci. 2014; 1:33–40. [Google Scholar]
- 4. UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hastings J., Owen G., Dekker A., Ennis M, Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C.. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016; 44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Placzek S., Schomburg I., Chang A., Jeske L., Ulbrich M., Tillack J., Schomburg D.. BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 2017; 45:D380–D388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schreiber F., Colmsee C., Czauderna T., Grafahrend-Belau E., Hartmann A., Junker A., Junker B.H., Klapperstück M., Scholz U., Weise S.. MetaCrop 2.0: managing and exploring information about crop plant metabolism. Nucleic Acids Res. 2012; 40:D1173–D1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar P., Han B.C., Shi Z., Jia J., Wang Y.P., Zhang Y.T., Liang L., Liu Q.F., Ji Z.L., Chen Y.Z.. Update of KDBI: Kinetic Data of Bio-molecular Interaction database. Nucleic Acids Res. 2009; 37:D636–D641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sayers E.W., Barrett T., Benson D.A., Bolton E., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Federhen S. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011; 39:D38–D51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gremse M., Chang A., Schomburg I., Grote A., Scheer M., Ebeling C., Schomburg D.. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011; 39:D507–D513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Courtot M., Juty N., Knüpfer C., Waltemath D., Zhukova A., Dräger A., Dumontier M., Finney A., Golebiewski M., Hastings J. et al. Controlled vocabularies and semantics in systems biology. Mol. Syst. Biol. 2011; 7:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000; 28:304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fleischmann A., Darsow M., Degtyarenko K., Fleischmann W., Boyce S., Axelsen K.B., Bairoch A., Schomburg D., Tipton K.F., Apweiler R.. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004; 32:D434–D437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S. et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016; 44:D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hucka M., Finney A., Sauro H.M., Bolouri H., Doyle J.C., Kitano H., Arkin A.P., Bornstein B.J., Bray D., Cornish-Bowden A. et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003; 19:524–531. [DOI] [PubMed] [Google Scholar]
- 18. Demir E., Cary M.P., Paley S., Fukuda K., Lemer C., Vastrik I., Wu G., D‘Eustachio P., Schaefer C., Luciano J. et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010; 28:935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruebenacker O., Moraru I.I., Schaff J.C., Blinov M.L.. Integrating BioPAX pathway knowledge with SBML. IET Syst. Biol. Models. 2009; 3:317–328. [DOI] [PubMed] [Google Scholar]
- 20. Funahashi A., Jouraku A., Matsuoka Y., Kitano H.. Integration of CellDesigner and SABIO-RK. In Silico Biol. 2007; 7:S81–S90. [PubMed] [Google Scholar]
- 21. Moraru I.I., Schaff J.C., Slepchenko B.M., Blinov M.L., Morgan F., Lakshminarayana A., Gao F., Li Y., Loew L.M.. Virtual Cell modelling and simulation software environment. IET Syst. Biol. 2008; 2:352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weidemann A., Richter S., Stein M., Sahle S., Gauges R., Gabdoulline R., Surovtsova I., Semmelrock N., Besson B., Rojas I. et al. SYCAMORE—a systems biology computational analysis and modeling research environment. Bioinformatics. 2008; 24:1463–1464. [DOI] [PubMed] [Google Scholar]
- 23. Dräger A., Zielinski D.C., Keller R., Rall M., Eichner J., Palsson B.O., Zell A.. SBMLsqueezer 2: context-sensitive creation of kinetic equations in biochemical networks. BMC Syst. Biol. 2015; 9:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Büchel F., Rodriguez N., Swainston N., Wrzodek C., Czauderna T., Keller R., Mittag F., Schubert M., Glont M., Golebiewski M. et al. Path2Models: large-scale generation of computational models from biochemical pathway maps. BMC Syst. Biol. 2013; 7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fuller J.C., Martinez M., Henrich S., Stank A., Richter S., Wade R.C.. LigDig: a web server for querying ligand-protein interactions. Bioinformatics. 2015; 31:1147–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wolstencroft K., Krebs O., Snoep J.L., Stanford N.J., Bacall F., Golebiewski M., Kuzyakiv R., Nguyen Q., Owen S., Soiland-Reyes S. et al. FAIRDOMHub: a repository and collaboration environment for sharing systems biology research. Nucleic Acids Res. 2017; 45:D404–D407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gaudet P., Michel P.A., Zahn-Zabal M., Britan A., Cusin I., Domagalski M., Duek P.D., Gateau A., Gleizes A., Hinard V. et al. The neXtProt knowledgebase on human proteins: 2017 update. Nucleic Acids Res. 2017; 45:D177–D182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gloaguen P., Bournais S., Alban C., Ravanel S., Seigneurin-Berny D., Matringe M., Tardif M., Kuntz M., Ferro M., Bruley C. et al. ChloroKB: a web application for the integration of knowledge related to chloroplast metabolic network. Plant Physiol. 2017; 174:922–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moretti S., Martin O., Van Du Tran T., Bridge A., Morgat A., Pagni M.. MetaNetX/MNXref—reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Res. 2016; 44:D523–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schomburg I., Jeske L., Ulbrich M., Placzek S., Chang A., Schomburg D.. The BRENDA enzyme information system-From a database to an expert system. J. Biotechnol. 2017; 261:194–206. [DOI] [PubMed] [Google Scholar]