Abstract
SMART (Simple Modular Architecture Research Tool) is a web resource (http://smart.embl.de) for the identification and annotation of protein domains and the analysis of protein domain architectures. SMART version 8 contains manually curated models for more than 1300 protein domains, with approximately 100 new models added since our last update article (1). The underlying protein databases were synchronized with UniProt (2), Ensembl (3) and STRING (4), doubling the total number of annotated domains and other protein features to more than 200 million. In its 20th year, the SMART analysis results pages have been streamlined again and its information sources have been updated. SMART’s vector based display engine has been extended to all protein schematics in SMART and rewritten to use the latest web technologies. The internal full text search engine has been redesigned and updated, resulting in greatly increased search speed.
INTRODUCTION
The concept of summarizing alignments of proteins or domains therein as profiles, collect them in a resource and scan new sequences against it has been already implemented in the 1980ies, e.g. using regular expressions (PROSITE release 1.0, (5)), alignment profiles, (ProfileScan, (6)) or property patterns (7) (ExCell, (8)). This precursor of SMART (Simple Modular Architecture Research Tool), a growing collection of profiles for shuffled extracellular domains, was in use as an inhouse tool soon after, i.e. in pre-WWW times (ExCell, (9,10)). With more data and increasing awareness and availability of mobile intracellular protein domains, the first version of SMART, released in 1997, aimed at creating a comprehensive resource for analyzing modular architecture of proteins; it used already then HMM profiles (11) and offered a web interface (12). Despite the focus shift in bioinformatics towards comparative genomics, transcriptomics and network biology, protein domain analysis remains an essential and important research tool, made easy by various frequently used online domain resources and databases, like Pfam (13), PANTHER (14) or PROSITE (15). Many of these, including SMART, are integrated into InterPro (16).
The SMART database (12) integrates manually curated hidden Markov models (11,17) for many domains with a powerful web-based interface offering various analysis and visualization tools. After 20 years since its inception, it remains a popular and widely used tool with close to 50 000 distinct users per month. In the following sections, we give an overview of the major developments and new features introduced since our last update (1).
EXPANDED DOMAIN COVERAGE
SMART was never intended to be exhaustive, and was initially focused on mobile domains. In order to provide context of other domains in modular proteins, but also to help in functional annotation, it continued to gradually expand its domain coverage with each new release. The current version introduces >100 new domains, compared to the last version (1), bringing the total to 1302. SMART’s domain annotation includes a significant amount of manual work and expertise, in particular in creating the high-quality underlying multiple sequence alignments and selecting the individual per-domain cut-off values. Other, more exhaustive databases, like Pfam (13), already annotated many of these domains, but SMART’s own manual annotation pipeline leads to partially different protein annotations, enabling increased hypothesis generation by biologists.
UPDATED PROTEIN DATABASES
The main underlying protein database in SMART combines of the complete Uniprot (2) with all stable Ensembl (3) proteomes. Current release contains >50 million proteins from around 460 thousand species, subspecies and strains. To minimize the impact of the inherently high redundancy of these databases, we use a per-species clustering method described in (18), which created 2.9 million multi protein clusters with a total of 5.5 million proteins.
In addition to the regular protein database described above, SMART offers a ‘genomic’ analysis mode that contains only proteins from completely sequenced genomes. Synchronized with the current STRING version 10.5 (4), it currently contains approximately 9.6 million proteins from 2031 complete genomes (238 Eukaryota, 1678 Bacteria and 115 Archaea).
NEW PROTEIN VISUALIZATION ENGINE AND UPDATED ANNOTATION PAGES
SMART version 8 introduces a vector based display engine for protein schematics (‘bubblograms’) throughout the server. Protein schematics in various list displays (such as domain architecture analysis results) are displayed as inline SVG (Scalable Vector Graphics) images, which seamlessly scale to the users display size, regardless of its resolution.
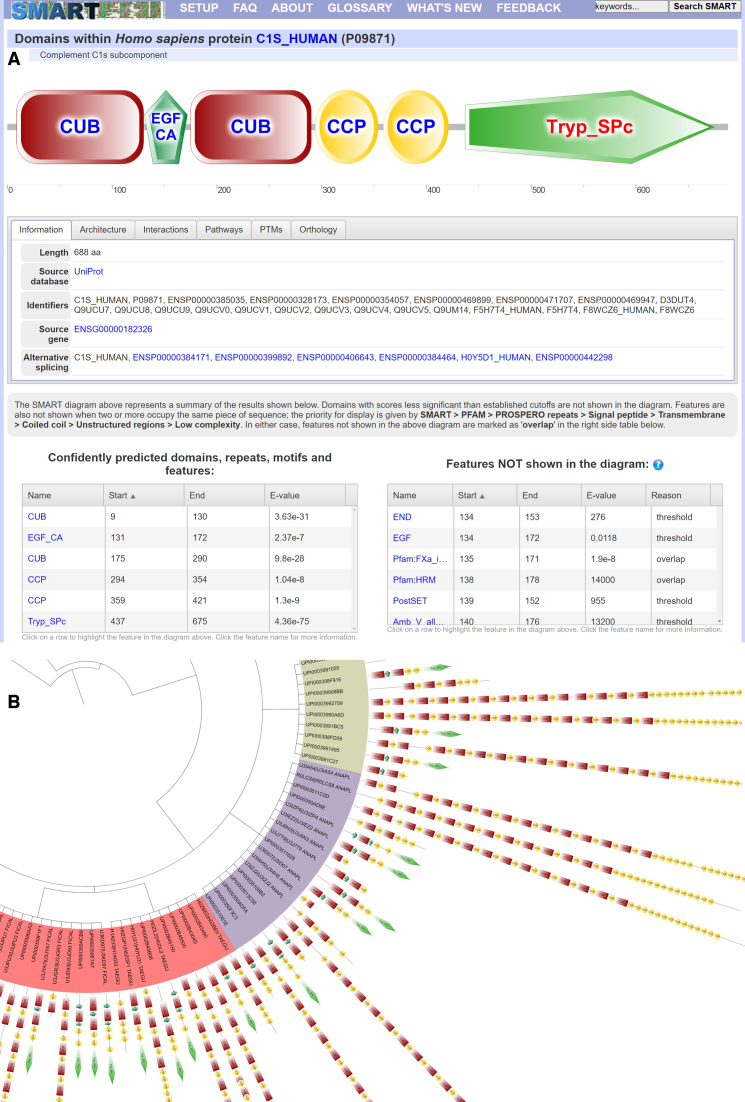
A vector based protein schematic display applet was used in the single protein annotation mode since the previous SMART version. However, it was implemented in Adobe Flash, which has been recently discontinued and will not be available in future web browsers. Therefore, we have implemented a new version based on HTML5 Canvas element, which should have a reasonable life time (Figure 1A). Schematics can be zoomed into any level without loss in display quality, as well as exported into vector or bitmap images at any resolution. A tool box within the interactive viewer provides access to several additional functions, for example allowing users to toggle the display of intron positions or to navigate among various alternative representations of proteins containing overlapping domain predictions.
Figure 1.
Example SMART protein annotation and tree export. (A) SMART annotation page for protein CS1_HUMAN. Protein schematic representations are displayed using vector graphics in a Canvas based applet. Schematics are zoomable without quality loss and exportable into high resolution SVG or bitmap images. Protein features selected in various data tables are dynamically highlighted directly in the viewer. Using the interactive scale, any protein region can be selected and submitted for further BLAST analysis. (B) An example domain architecture analysis result, exported from SMART directly into the interactive Tree Of Life (iTOL) version 3 (21).
The new protein viewer interactively ties various parts of the annotation page. Selecting a predicted domain or other feature in any of the data tables will automatically highlight its position in the protein. Since many predicted features are not directly displayed in the protein schematic (mostly due to overlaps), this function simplifies the visual identification of relations among different protein features.
Various parts of the protein sequence can be interactively selected independent of the annotated features, and submitted to further BLAST analysis, when a more fine-grained evaluation is required.
Detailed information about any detected protein feature can be displayed in streamlined floating popup dialogs, enhancing the user experience and lowering the need to navigate across different web pages. Condensed version of domain annotation pages is included in the dialogs, with optional links to the complete annotation. In addition, several convenience functions are included, allowing users to copy the underlying amino acid sequence to their clipboard, or to submit the subsequence for further BLAST analysis.
UPDATED EXTERNAL INFORMATION SOURCES
Protein orthology data are parsed from the the eggNOG database version 4.5 (19) and covers ∼7.5 million proteins from >3500 species. SMART’s annotation pages show a detailed list of all orthologous groups that include the protein annotated, with their description and taxonomic class. Cross links to eggNOG are provided, with detailed overviews of each orthologous group as well as the associated alignments and phylogenetic trees.
Data on posttranslational protein modifications, which are displayed since the last SMART release, have been synchronized with the latest version 2 of the PTMcode database (20). SMART displays the total numbers of various posttranslational modifications annotated in a particular protein, with links to the detailed annotation pages in PTMcode, where users can explore the modifications and their possible functional associations within the protein, as well as with their direct interaction partners.
EXPANDED PROTEIN INTERACTION DATA
With the update of the underlying protein databases, we have also synchronized our protein interaction data with the version 10.5 of the STRING database (4). Updated graphical representations of putative interaction partners are now available for >9.5 million proteins.
UPDATED TAXONOMIC TREE DATA EXPORT
Domain architecture analysis functions in SMART allow users to simply access proteins containing combinations of particular domains. These can be also generated using combinations of GO terms associated to protein domains, and restricted to various taxonomic classes. In addition to the standard SMART protein schematic visualization, these data can also be exported into FASTA files or phylogenetic trees. The phylogenetic tree export has been completely rewritten and made compatible with the version 3 of the Interactive Tree of Life (iTOL) (21), with which these trees and their associated protein domain datasets can be further annotated (Figure 1B). Furthermore, backend taxonomic information used for the tree generation was synchronized with the latest NCBI taxonomy database.
BACKEND OPTIMIZATIONS AND EXPANDED SEARCH ENGINE
The backend of SMART is a relational database management system (RDBMS), powered by the PostgreSQL engine, which stores the annotation of all SMART domains, protein annotation and sequences, taxonomy information and the pre-calculated protein analyses for the entire Uniprot (2), Ensembl (3) and STRING (4) proteomes. In addition to the predictions of all SMART and Pfam domains, this includes various protein intrinsic features, like signal peptides, transmembrane and coiled coil regions. Due to constant growth of the number of annotated features, we are regularly restructuring our backend databases, and optimizing various parts of the server code in order to make the user experience satisfactory. Additionally, the server hardware that powers the sequence annotation searches and database queries has been replaced and significantly expanded with additional RAM and CPUs, greatly increasing the processing speed of user submitted proteins, and lowering the overall response times.
SMART’s full text search engine allows users to quickly identify domains or proteins based on their annotation and other associated text. The current version introduces an updated search backend, providing access to a wider array of text information associated with each protein/domain, while offering increased search speed.
CONCLUSION
Since the initial release of SMART more than 20 years ago, our goal has been to provide a useful biological web resource, characterized by a high quality of underlying data and a powerful, simple user interface, even in the context of a very low funding level (on average less than 1 FTE over the last 10 years). Thus, we are confident that we are able to continue to modestly expand our coverage, keep in line with latest web standards and implement new features to make using SMART a better and more enjoyable experience to both existing and new users.
ACKNOWLEDGEMENTS
We would like to thank our colleague Tobias Doerks, who has recently passed away, for his numerous contributions, suggestions and expertise, which will remain integrated in various aspects of SMART. Furthermore, members of the Bork group are acknowledged for valuable discussions.
FUNDING
German Network for Bioinformatics Infrastructure (de.NBI); EMBL. Funding for open access charge: European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1. Letunic I., Doerks T., Bork P.. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015; 43:D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. The UniProt, C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al. Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991; 19(Suppl):2241–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gribskov M., McLachlan A.D., Eisenberg D.. Profile analysis: detection of distantly related proteins. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:4355–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bork P., Grunwald C.. A method for property pattern searches in protein sequence databases, demonstrated by detection of GTP binding sites. Stud. Biophys. 1989; 129:231–240. [Google Scholar]
- 8. Bork P. Recognition of functional regions in primary structures using a set of property patterns. FEBS Lett. 1989; 257:191–195. [DOI] [PubMed] [Google Scholar]
- 9. Bork P. Shuffled domains in extracellular proteins. FEBS Lett. 1991; 286:47–54. [DOI] [PubMed] [Google Scholar]
- 10. Bork P., Ouzounis C., Sander C., Scharf M., Schneider R., Sonnhammer E.. Comprehensive sequence analysis of the 182 predicted open reading frames of yeast chromosome III. Protein Sci. 1992; 1:1677–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krogh A., Brown M., Mian I.S., Sjolander K., Haussler D.. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 1994; 235:1501–1531. [DOI] [PubMed] [Google Scholar]
- 12. Schultz J., Milpetz F., Bork P., Ponting C.P.. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D.. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017; 45:D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sigrist C.J., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I.. New and continuing developments at PROSITE. Nucleic Acids Res. 2013; 41:D344–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.Y., Dosztanyi Z., El-Gebali S., Fraser M. et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017; 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Letunic I., Doerks T., Bork P.. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009; 37:D229–D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M.C., Rattei T., Mende D.R., Sunagawa S., Kuhn M. et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016; 44:D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minguez P., Letunic I., Parca L., Garcia-Alonso L., Dopazo J., Huerta-Cepas J., Bork P.. PTMcode v2: a resource for functional associations of post-translational modifications within and between proteins. Nucleic Acids Res. 2015; 43:D494–D502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Letunic I., Bork P.. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016; 44:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]