Abstract
Real-time quantitative polymerase chain reaction (qPCR) is one of the most important methods for analyzing the expression patterns of target genes. However, successful qPCR experiments rely heavily on the use of high-quality primers. Various qPCR primer databases have been developed to address this issue, but these databases target only a few important organisms. Here, we developed the qPrimerDB database, founded on an automatic gene-specific qPCR primer design and thermodynamics-based validation workflow. The qPrimerDB database is the most comprehensive qPCR primer database available to date, with a web front-end providing gene-specific and pre-computed primer pairs across 147 important organisms, including human, mouse, zebrafish, yeast, thale cress, rice and maize. In this database, we provide 3331426 of the best primer pairs for each gene, based on primer pair coverage, as well as 47760359 alternative gene-specific primer pairs, which can be conveniently batch downloaded. The specificity and efficiency was validated for qPCR primer pairs for 66 randomly selected genes, in six different organisms, through qPCR assays and gel electrophoresis. The qPrimerDB database represents a valuable, timesaving resource for gene expression analysis. This resource, which will be routinely updated, is publically accessible at http://biodb.swu.edu.cn/qprimerdb.
INTRODUCTION
Real-time quantitative polymerase chain reaction (qPCR) is one of the most powerful and effective research tools currently available for molecular genetic studies. This tool is widely used to quantitatively analyze gene expression levels and to identify differentially expressed genes under various experimental treatments, even those with low expression levels (1). This method is also used for effective single nucleotide polymorphism (SNP) genotyping, genetically modified organism (GMO) and pathogen detection, and human in vitro diagnostics. However, the accuracy and consistency of qPCR detection strongly depends on the specificity and efficiency of the designed primers and on various experimental factors, leading to considerable variation in qPCR assay results and, at times, even incorrect conclusions. Moreover, the transcription levels vary greatly among different gene regions, as observed in several eukaryotes (e.g. human, mouse and chicken) (2–4). Thus, gene expression analysis using primers designed based on various regions of a gene may yield different results. Furthermore, qPCR primer design is a lengthy process, since homologous sequence alignment, primer design and confirmation of the specificity and amplification conditions are time-consuming processes, and designing primers for polyploid organisms demands even more time.
To address the abovementioned challenges, various computational methods, programs and databases have been developed. Primer3 (5), an important program for primer design, is a popular tool that forms the basis for many other tools and is often used for high-throughput genomics applications. Several local or web-based programs based on Primer3 have been developed for batch primer design, including BatchPrimer3 (6), QuantPrime (7), PCRTiler (8), PRIMEGENS (9) and PrimerMapper (10). For most of these programs, primer specificity is only evaluated using the Nucleotide Basic Local Alignment Search Tool (BLASTN) or sequence similarity searching; however, other essential factors necessary for successful qPCR should also be considered.
The thermodynamics-based specificity-checking program, MFPrimer-2.0 (11) and the MapReduce-based method, MRPrimer (12), were recently developed to overcome the drawbacks of the abovementioned homology testing methods. Although these programs have been optimized, it is still difficult for most researchers to design many qPCR primers that simultaneously satisfy the requirements for stringent, uniform amplification conditions. Therefore, various qPCR primer databases have been developed to provide pre-designed, specific qPCR primers, such as AtRTPrimer (13), GETPrime 2.0 (14), MRPrimerW (15), PrimerBank (16), qPrimerDepot (17) and RTPrimerDB (18). Unfortunately, these databases were designed based on only a few important organisms. For example, AtRTPrimer is a primer database for thale cress (Arabidopsis thaliana), both PrimerBank and MRPrimerW contain primers for human (Homo sapiens) and mouse (Mus musculus) and GETPrime 2.0 includes gene- and transcript-specific qPCR primers for 13 species (13–16).
Recent innovations in next-generation sequencing technology and the rapidly declining cost of sequencing have increased the number of genome sequences available, especially for agriculturally important livestock and plants. However, gene-specific qPCR primers have not yet been designed for most of these recently sequenced genomes and are therefore not included in any comprehensive database.
In the current study, to provide a comprehensive, uniform platform to researchers and to include as many sequenced genomes as possible, we developed a qPCR primer design workflow and generated large numbers of qPCR primer pairs spanning 147 important organisms. Based on these data, we developed the qPrimerDB database, which systematically integrates large numbers of primer pairs for each gene, as well as alternative gene-specific primers. The qPrimerDB database enables both the batch downloading and rapid retrieval of qPCR primers for selected genes, via a user-friendly interface. We validated the quality of the qPCR primers through qPCR analysis of 66 randomly selected genes. This qPrimerDB represents an important database of qPCR primer sets that can be conveniently used for qPCR analysis.
MATERIALS AND METHODS
Data sources
qPrimerDB has collected the whole-genome sequences of 147 important organisms (including 80 animals, 66 plants and one fungus), as well as the corresponding coding/mRNA sequences (Supplementary Table S1). Of the 147 genome sequences included, genome data from A. thaliana were downloaded from The Arabidopsis Information Resource (http://www.arabidopsis.org/) (19), 33 plant genomes were downloaded from Phytozome 12.0 (http://www.phytozome.net) (20), four legume genomes were obtained from Legume Information System (https://legumeinfo.org) (21), two plant genomes were acquired from PLAZA 3.0 (http://bioinformatics.psb.ugent.be/plaza) (22), and the 107 remaining genomes were retrieved from Ensembl (http://www.ensembl.org/) (23).
Primer generation
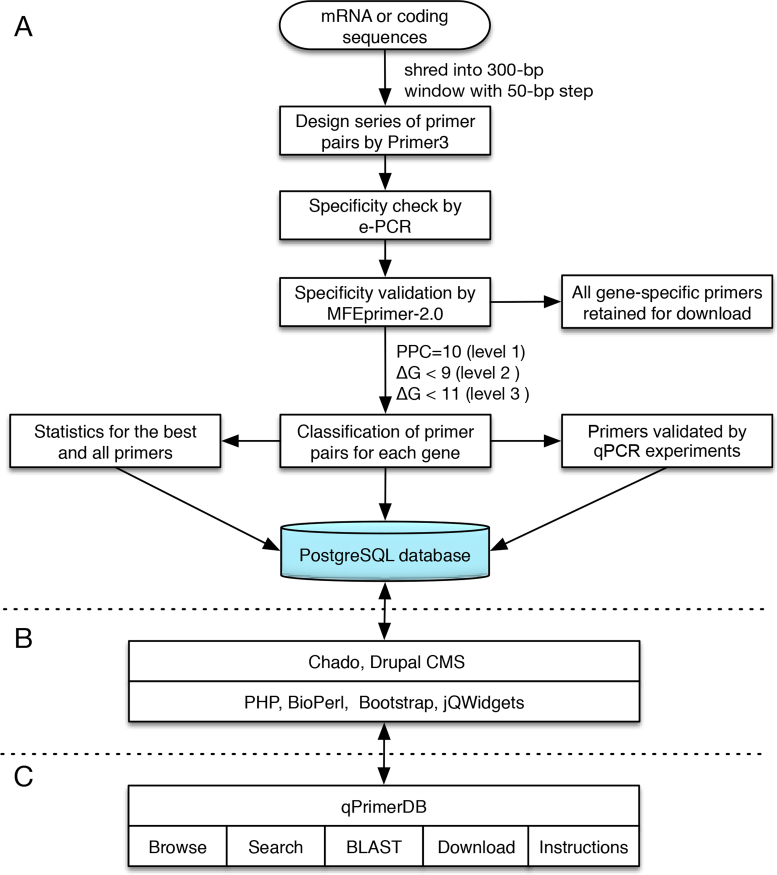
The workflow for qPCR primer generation in the qPrimerDB integrates several existing programs, including BBMap (https://sourceforge.net/projects/bbmap), Primer3 (5), electronic PCR (e-PCR) (24), MFPrimer-2.0 (11) and BLAT (25), using in-house shell scripts, allowing automatic high-throughput primer pair design, filtering and classification to be performed based on the level of specificity (Figure 1A). There are seven steps in the qPrimerDB workflow, with the following functions.
Figure 1.
Workflow for the development of qPrimerDB. (A) Workflow for the generation and classification of gene-specific qPCR primer pairs. The best and all primer sequences are loaded into the PostgreSQL database. (B) Implementation of qPrimerDB via the integration of different programs. (C) Organization of qPrimerDB.
Step 1 (sequence pre-processing): removes all duplicated coding/mRNA sequences, leaving the longest coding/mRNA sequence for each gene. After treatment, 3 598 514 representative coding/mRNA sequences, spanning the 147 eukaryotic organisms, were obtained (Supplementary Table S1).
Step 2 (sequence fragmentation): designs as many qPCR primers as possible for each gene. All sequences retained in Step 1 are shredded into a series of short, overlapping template fragments using BBMap, with the specified window size and step size currently set to 300 and 50 bp, respectively. The window and step size can be adjusted for organisms with complex genomes, but this will dramatically increase computation time.
Step 3 (generation of unique candidate primer pairs): performs automatic primer design for all of the shredded template fragments using Primer3 with the following parameters: amplicon size 80–300 bp; amplicon GC content 40–60%, with an optimal GC content of 50%; primer length 18–28 nt, with an optimal length of 22 nt; and melting temperature (Tm) 58–64°C, with an optimal Tm of 60°C and maximum Tm difference, per primer pair, of less than 3°C. For each gene, duplicated candidate primer pairs generated at neighboring windows are removed.
Step 4 (specificity checking by ePCR): employs ePCR (24) to analyze the specificity of all designed primers. All primers with one mismatch or indel against any off-target sequence are filtered out. The remaining candidate primer pairs are used for the next round of specificity validation.
Step 5 (specificity checking using MFEprimer-2.0): indexes the databases for the coding/mRNA sequence of the target organism, based on the k-mer index algorithm, using the thermodynamics-based gene-specificity checking program, MFEprimer-2.0 (11). By default, the k-mer is set to 9, which is sufficient for primer specificity checking; a lower value can be set if more stringent conditions are desired, but this will increase computational time. Specificity of qPCR primer pairs against the genomic sequence and coding/mRNA sequence databases is then estimated, and related parameters, such as the binding site in the sequence, the size and GC content of the amplicon, and the Tm and Gibbs free energy (ΔG) of the forward and reverse primers can be recorded.
Step 6 (classification of primer pairs): primer pairs for each gene are divided into three levels, based on primer pair coverage (PPC) and the binding stability of the binding site, ΔG, as follows: (i) level 1: the primer pairs will be removed if their PPCs against any non-specific amplicons are higher than 10; (ii) level 2: the primer pairs will be filtered out if the ΔG of binding for either the forward or reverse primer to off-target sequences is less than −9 kcal/mol; (iii) level 3: similar to level 2, except that the ΔG is set to −11 kcal/mol. In addition, all qualified primer pairs are further divided into two categories: the best primers and alternative primers. The former category includes only one best, unique primer pair with the lowest total ΔG of the pair, which is the gene-specific qPCR primer pair recommended for the detection of gene expression. The latter category includes all (both the best and alternative) gene-specific qPCR primer pairs.
Step 7 (obtain amplicon parameters): conducts a BLAT search using each pair of primers against the target genomic sequence. The number of exons spanned and the genomic position of each primer and amplicon can be determined to select primers spanning an exon-intron boundary, and this information can be used to avoid amplifying contaminating genomic DNA.
After computation, the workflow generated a total of 51 091 785 gene-specific qPCR primer pairs, corresponding to 93.4% of the average coverage ratios, per organism, and an average of 15.3 primer pairs per gene (Table 1 and Supplementary Table S1). In human and mouse, more than 19 and 22 primer pairs, per gene, were obtained, with a coverage ratio of 78.8 and 90.3%, respectively (Supplementary Table S1), indicating that the results of our workflow are in accordance with those in MRPrimerW (15) and PrimerBank (16). Moreover, it is more difficult to design qPCR primers for plants than for animals. Using the same workflow, the coverage ratios decreased from 93.80% in animals to 93.08% in plants, and the number of primer pairs per gene significantly decreased from 17.20 to 12.55 (permutation test P-value < 0.001). This reduction might have been due to the difficulty in designing qPCR primers for polyploid plants, such as oilseed rape (Brassica napus), soybean (Glycine max), potato (Solanum tuberosum) and tomato (Solanum lycopersicum). Hence, this database would greatly facilitate qPCR experiments for studies based on organisms with complex genomes.
Table 1. Comparison between qPrimerDB and previously established qPCR primer databases.
| Database | Number of organisms | Total number of genes | Number of genes covered | Total number of primer pairs | The best primer recommendation | Number of exons spanned | Batch download | Amplicon and cDNA sequences | BLAST for ID conversion | Primer melting temperature | Gibbs free energy of primer |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PrimerBank | 2 | 61 425 | 306 800 | × | × | × | √ | × | √ | × | |
| MRPrimerW | 2 | 56 227 | 56 173 | 341 963 135 | √ | × | √ | × | × | √ | × |
| RTPrimerDB | 21 | 5686 | 7963 | × | × | × | × | × | √ | × | |
| GETPrime 2.0 | 13 | 144 410 | 134 868 | 1 175 874 | √ | × | √ | × | × | √ | × |
| qPrimerDB | 147 | 3 598 514 | 3 331 426 | 51 091 785 | √ | √ | √ | √ | √ | √ | √ |
Primer validation and comparison
To validate amplification specificity and to check for the presence of non-specific amplicons, qPCR assays were performed using 66 randomly selected genes from thale cress, oilseed rape, rice (Oryza sativa), sweet orange (Citrus sinensis), silkworm (Bombyx mori) and zebrafish (Danio rerio). In these qPCR experiments, only the best primer pair for each gene from qPrimerDB was selected, as summarized in Supplementary Table S2. To ensure the reliability of experimental results, the setup and validation of qPCR were performed, as prescribed in the MIQE guidelines (26). Based on the results of melting curve analysis and gel electrophoresis, all selected primer pairs were demonstrated to be of high specificity and amplification efficiency (Supplementary Figure S1).
In addition, to test the specificity and accuracy of the primer design workflow used in qPrimerDB, we collected 454 and 137 qPCR primer pairs for A. thaliana and B. mori, respectively, from 55 previously published studies (Supplementary Table S3). Using the validation and classification steps in the qPrimerDB workflow, the specificity and classification of all collected primers were determined. The results showed that the ratios of gene-specific primers were 75.87% for A. thaliana and 98.23% for B. mori in qPrimerDB, whereas values collected from the published studies were only 71.09 and 79.65%, respectively (Supplementary Table S4). These assays established that the automatic primer design workflow developed in qPrimerDB generated high quality gene-specific qPCR primers, whose specificity and consistency were superior to those of manually designed primers.
We also compared the qPCR primers for H. sapiens generated with MRPrimer and qPrimerDB workflows. For both two methods, only the best pre-commutated primers were retained and used for specificity and classification comparison. The results showed that a total of 14 500 MRPrimer and 17 632 qPrimerDB qPCR primers were gene-specific, and the ratio of level 1 primer in qPrimerDB was 50.36%, much higher than that in MRPrimer (13.14%) (Supplementary Table S5), indicating that the qPrimerDB workflow might well be one of the most effective gene-specific qPCR primer design tools, suitable for automatic primer design for newly sequenced genomes.
Database implementation
The qPrimerDB is implemented using PostgreSQL 9.6 (http://www.postgresql.org), PHP 5.5 (http://www.php.net), Apache Web Server 2.4 (http://www.apache.org) and BioPerl 1.6 (http://bioperl.org) on a Linux CentOS 6.8 operating system (Figure 1B). Our database is supported by integration with the Chado (27) database schema and Drupal (28), a popular Content Management System. As described for SilkPathDB (29), the qPrimerDB architecture is composed of four layers. The core and inner most layer is the Chado schema, which stores and organizes all qPrimerDB data, and is managed with the PostgreSQL. The second layer is a configurable layer that controls the interactions between the core layer and the third layer, programs of which process and respond to queries from the outermost layer, the web interfaces presenting data to users and forms the querying database. To provide a friendly interface for users accessing the database from desktop and mobile devices, the Bootstrap framework (http://getboot strap.com) is employed. PHP and BioPerl scripts are used to generate primer index and search results by retrieving primer information for query genes and organisms inputted by the user. The BLAST database is built with NCBI BLAST+ 2.6.0 (30), with the multiple database search function. Finally, jQWidgets (http://www.jqwidgets.com) is used to organize and present the feature index, search, and BLAST results.
RESULTS
Database organization
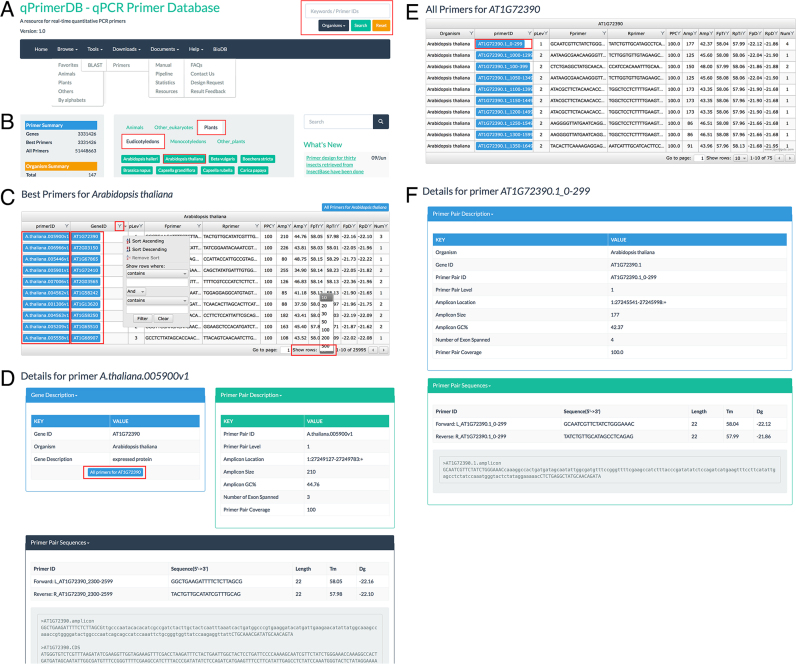
The qPrimerDB website comprises seven functional sections, including ‘Home’, ‘Browse’, ‘Tools’, ‘Download’, ‘Documents’, ‘Help’ and ‘Search’ (Figures 1C and 2A). From the ‘Home’ page, users can view the ‘Introduction’, update news and statistics for the qPrimerDB database. A convenient browse function is also provided on the ‘Home’ page. In the ‘Browse’ section, we classify all 147 organisms into ‘Favorites’, ‘Animals’, ‘Plants’ and ‘Others’, providing an easy way to find the target organism. We also provide a page that sorts the 147 organisms by scientific name in alphabetical order. In the ‘Tools’ section, we embedded the BLAST program, which can be used to allow identifiers to be converted between different databases. In the ‘Download’ section, qPrimerDB offers a user-friendly interface for users to conveniently batch download the best or all primer sequences for the target organism. By selecting ‘Documents’ in the navigation bar, users can find the manual, pipelines for primer design and database implementation, statistics for each organism and related resources. On the ‘Help’ page, users can view frequently asked questions (FAQs) and answers. Users can also submit their qPCR results and design requests via three different feedback pages. Our database will be updated regularly based on user feedback. A search tool is available on the upper right corner of every qPrimerDB page. Using the search box, users can search for primer pairs by choosing single or multiple organisms and inputting gene name(s), primer IDs, or keywords of interest and export detailed information for their target genes.
Figure 2.
Screenshots of the navigation bar and browse module in qPrimerDB. (A) Navigation bar of qPrimerDB. A search box is provided in the upper-right corner of every page to enable convenient searching of keywords of interest. (B) Browse interface on the ‘Home’ page. Users can browse the organism of interest, for example, by sequentially clicking ‘Plants’, ‘Eudicotyledons’ and ‘Arabidopsis thaliana’. (C) Example of A. thaliana primers in table format. Both the record number per page and the order of each column can be adjusted, as needed. (D) qPrimerDB primer details page. Detailed information for primer ID A. thaliana.005900.v1 is presented in three sections: Gene Description (gene ID, organism, gene description, and a blue button ‘All primers for AT1G72390’); Primer Pair Description (primer pair ID and level, amplicon location, amplicon size, amplicon GC content, number of exons spanned, PPC); and Primer Pair Sequences (Tm values of primers, primer sequences, primer length and amplicon and template coding/mRNA sequences). (E) Example of all primers for AT1G72390 in table format. After clicking the blue button in Figure 2D, all primers will be listed in table format. (F) Detailed information for primer ID AT1G72390.1_0–299 is presented in two sections: Primer Pair Description (organism, Gene ID, primer pair ID and level, amplicon location, amplicon size, amplicon GC content, number of exons spanned, PPC); and Primer Pair Sequences (Tm values of primers, primer sequences, primer length and amplicon sequence).
Web interface and usage
Browse function
In our database, the 147 organisms are organized by taxonomic group and in alphabetical order. To increase browsing efficiency, the 147 organisms were initially classified into three categories: animals, plants and other eukaryotes. The 80 animals were further divided into groups including four birds and reptiles, 10 fishes, 17 insects, 37 mammals and 12 other animals, while the 66 plants were separated into groups including 39 eudicotyledons, 18 monocotyledons, and nine other plants. In addition, the 147 organisms were sorted by alphabetical order based on scientific name. There are two ways to browse all the information pertaining to qPCR primer pairs for a target organism. We use the model plant ‘A. thaliana’ as an example to demonstrate the browse function in qPrimerDB. Using the first method, users can browse information about the organism of interest by selecting its classification and scientific name from the ‘Home’ page. After sequentially clicking ‘Plants’, ‘Eudicotyledons’ and ‘A. thaliana’ (Figure 2B), the ‘Results’ page displays detailed information about the best primer pairs for 25 995 genes, including primer ID, gene ID, primer level, forward primer sequence, reverse primer sequence, PPC, amplicon size, amplicon GC content, Tm of forward primer, Tm of reverse primer, ΔG of forward primer, ΔG of reverse primer and number of exons spanned. The 25995 records are displayed in 2600 pages with 10 entries per page (by default), which could be adjusted by changing the ‘Show row’ to 20, 30, 50, 100, 200 or 500 (Figure 2C). The order of values displayed in each column of the ‘Results’ table can also be changed by choosing ‘Sort Ascending’ or ‘Sort Descending’, and keywords in a column can be searched in different ways by choosing terms, such as ‘contains’, ‘does not contain’ and ‘equal’, and so on.
More detailed information can be accessed for each gene via the hyperlink in the blue rectangle in the primer ID and gene ID columns. For example, users can view the detailed information for best primer of AT1G72390 by clicking the hyperlink of primer ID A.thaliana.005900.v1, including gene description, primer pair description, hyperlink for ‘All primers for AT1G72390’ and amplicon and template sequences (Figure 2D). The table of all primers for AT1G72390 (Figure 2E) could be viewed by clicking the hyperlinks ‘AT1G72390’ in ‘Best Primers for A. thaliana’ page (Figure 2C) or ‘All primers for AT1G72390’ in ‘Details for primer A.thaliana.005900.v1’ page (Figure 2D). Then, detailed information for each of all primers (Figure 2F) could be viewed by clicking the primer ID of interest. Using the second method, users can browse the organism of interest by selecting ‘Browse’ in the navigation bar, followed by ‘A. thaliana’ from the ‘Favorites’, ‘Plants’ or ‘By alphabets’ page. The remaining browsing steps are performed as described for the first method.
Search tools
To search for genes of interest, we supplied two search tools: a search box on the top right corner of every page and an embedded BLAST server. When using the search box, users should select single or multiple target organisms by sequentially clicking the ‘Organisms’ button and checking the box next to the scientific name(s) and then inputting the gene description, gene ID or primer ID in the search box (Supplementary Figure S2A). After clicking the ‘Search’ button, the results, which are presented in table format, can be sorted and filtered as a ‘Results’ table in the ‘Browse’ page (Supplementary Figure S2B). This tool also allows users to select and export search results of interest as XLS or JSON files by clicking the check boxes located next to the target primers. Given the different identifiers and symbols used for the same gene, across different databases, we have provided a BLAST similarity search server for transferring gene IDs that are unavailable to qPrimerDB to available gene IDs. Thereafter, users can perform BLAST searches against single or multiple organisms of interest using their own gene sequences in FASTA format (Supplementary Figure S2C). If the output format is set to table format, users can obtain detailed information about primers of interest in the ‘Results’ page via a hyperlink under Subject heading (Supplementary Figure S2D).
Batch download tools
In qPrimerDB, all qPCR primers and the best primers designed for each organism are separately compressed into two zip files. Users can download these datasets in bulk via ‘Datasets’, a hyperlink in the ‘Downloads’ drop-down menu in the navigation bar (Figure 2A). After downloading and decompression of the two zip files containing all qPCR primers or the best primers, two more columns (forward and reverse primer IDs), in addition to the results in the browse and search tools, can be viewed. Thereafter, users can select multiple qPCR primers for the genes of interest and submit them for oligo synthesis, without manually adding any further information.
Primer statistics
Users can view the statistics for the primers for each genome in qPrimerDB by clicking ‘Statistics’ in the ‘Documents’ drop-down menu (Figure 2A). We summarized the unique gene number, number of best primers in each of three levels and total number of primers, sum of the best primers and all primers, percentage of genes with best primersand average number of primer pairs for each of the 147 organisms.
Comments
We provide the user with three feedback forms. Both ‘Design Request’ and ‘Results Feedback’ can be selected from the ‘Help’ drop-down menu in the navigation bar (Figure 2A). In the Design Request form, users can send us design requests for organisms not included in our database by providing the download links for unique coding/mRNA, protein and genome sequences. To increase the running efficiency, we also welcome links to annotation files. In the latter form, users are invited to submit their qPCR detection results (from melting curve analysis and/or gel electrophoresis) to us, especially the specificity of the experimentally examined primer pairs. Such information will be used for further improving qPrimerDB, as well as by other researchers with the same interests. Reliable primer pairs validated by qPCR will be marked in the database and will be strongly recommended, and non-specific primer pairs will be removed in subsequent versions of qPrimerDB. We also provide a ‘Contact Us’ tab in the ‘Help’ menu, where users can submit comments or suggestions about the content and structure of the database.
Manual and FAQs
We generated a user’s manual that will allow users to easily understand the features of our database, and how to obtain primer pairs and related information of interest easily and efficiently. A step-by-step guide for the Browse, Search and Download tools can be found under the ‘Manual’ tab in the ‘Documents’ menu (Figure 2A). Some FAQs and corresponding answers can be viewed by clicking the ‘FAQs’ tab in the ‘Help’ menu, including the introduction and usage of qPrimerDB. This information will help users efficiently find information in the database.
DISCUSSION AND FUTURE PROSPECTS
Numerous computer programs have been developed to facilitate the automation of qPCR primer design, many of which are widely used (5–12). In addition, several databases containing pre-computed or experimentally validated qPCR primers based on different programs or analysis workflows have been developed (13–18). These programs and databases help improve the efficiency and accuracy of qPCR assays. Although numerous genomes of important organisms have been sequenced to date, only a few (such as human, mouse and thale cress) were included in previously released primer databases. Therefore, there is an urgent need to develop a platform to design and validate gene-specific qPCR primers for a broader spectrum of genomes with the same primer filtering constraints, via a uniform design workflow. To this end, the qPrimerDB database was developed based on a thermodynamics-based design and validation workflow. A comparison between the thermodynamics-based qPrimerDB and the MapReduce-based MRPrimer indicated that qPrimerDB workflow might represent one of the most effective gene-specific qPCR primer design programs. The comparison between primers collected from previously published studies and the corresponding primers in qPrimerDB provided further support for the specificity and consistency of qPCR primers included in qPrimerDB.
To the best of our knowledge, qPrimerDB is the most comprehensive qPCR primer database available to date, and incorporates information for 147 organisms, including 80 animals, 66 plants and 1 fungus. We believe that this valuable resource will assist the research community by enabling the efficient and specific performance of qPCR experiments. We plan to update the database periodically and incorporate information for additional organisms after running the analysis workflow. For example, we recently designed primers for 60 additional, important organisms (such as upland cotton (Gossypium hirsutum), wheat (Triticum aestivum) and cabbage (Brassica oleracea var. capitata)). We also plan to include more types of qPCR primers in our database, such as those used for microRNA analysis, internal controls and detection of genetically modified organisms. Gene-specific primers that have been experimentally validated will be marked in the database, based on user feedback, and will be strongly recommended to users, while non-specific primers will be removed from the database.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank William Lucas, Distinguished Professor of University of California, Davis, Dr Kathleen Farquharson and Dr Jennifer A. Lockhart for their critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Plan [2016YFD0101007 to K.L.]; National Program on Key Basic Research Project [2015CB150201 to W.Q.]; National Natural Science Foundation of China [31571701 to K.L., U1302266 to J.L.]; National High Technology Research and Development Programs of China [2013AA102602 to K.L]; Programme of Introducing Talents of Discipline to Universities [B12006 to J.L.]; Key Special Program of China National Tobacco Corporation [TS-02-20110014 to B.L.]. Funding for open access charge: Southwest University, China [XDJK2012A009 to K.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Holland M.J. Transcript abundance in yeast varies over six orders of magnitude. J. Biol. Chem. 2002; 277:14363–14366. [DOI] [PubMed] [Google Scholar]
- 2. Arhondakis S., Clay O., Bernardi G.. GC level and expression of human coding sequences. Biochem. Biophys. Res. Commun. 2008; 367:542–545. [DOI] [PubMed] [Google Scholar]
- 3. Sémon M., Mouchiroud D., Duret L.. Relationship between gene expression and GC-content in mammals: Statistical significance and biological relevance. Hum. Mol. Genet. 2005; 14:421–427. [DOI] [PubMed] [Google Scholar]
- 4. Rao Y.S., Chai X.W., Wang Z.F., Nie Q.H., Zhang X.Q.. Impact of GC content on gene expression pattern in chicken. Genet. Sel. Evol. 2013; 45:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G.. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012; 40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. You F.M., Huo N., Gu Y., Luo M., Ma Y., Hane D., Lazo G.R., Dvorak J., Anderson O.D.. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008; 9:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arvidsson S., Kwasniewski M., Riano-Pachon D.M., Mueller-Roeber B.. QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics. 2008; 9:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gervais A.L., Marques M., Gaudreau L.. PCRTiler: automated design of tiled and specific PCR primer pairs. Nucleic Acids Res. 2010; 38:W308–W312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kushwaha G., Srivastava G.P., Xu D.. PRIMEGENSw3: a web-based tool for high-throughput primer and probe design. Methods Mol. Biol. 2015; 1275:181–199. [DOI] [PubMed] [Google Scholar]
- 10. O’Halloran D.M. PrimerMapper: high throughput primer design and graphical assembly for PCR and SNP detection. Sci. Rep. 2016; 6:20631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qu W., Zhou Y., Zhang Y., Lu Y., Wang X., Zhao D., Yang Y., Zhang C.. MFEprimer-2.0: a fast thermodynamics-based program for checking PCR primer specificity. Nucleic Acids Res. 2012; 40:205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim H., Kang N.N., Chon K.W., Kim S., Lee N.H., Koo J.H., Kim M.S.. MRPrimer: a MapReduce-based method for the thorough design of valid and ranked primers for PCR. Nucleic Acids Res. 2015; 43:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han S., Kim D., Holland M., Horak C., Snyder M., Steve R., Helen J., Pattyn F., Speleman F., De Paepe A. et al. AtRTPrimer: database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC Bioinformatics. 2006; 7:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. David F.P.A., Rougemont J., Deplancke B.. GETPrime 2.0: gene- and transcript-specific qPCR primers for 13 species including polymorphisms. Nucleic Acids Res. 2017; 45:D56–D60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim H., Kang N., An K., Koo J., Kim M.-S.. MRPrimerW: a tool for rapid design of valid high-quality primers for multiple target qPCR experiments. Nucleic Acids Res. 2016; 44:W259–W266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X., Spandidos A., Wang H., Seed B.. PrimerBank: A PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012; 40:D1144–D1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui W., Taub D.D., Gardner K.. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007; 35:D805–D809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lefever S., Vandesompele J., Speleman F., Pattyn F.. RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res. 2009; 37:D942–D945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M. et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012; 40:D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gonzales M.D., Archuleta E., Farmer A., Gajendran K., Grant D., Shoemaker R., Beavis W.D., Waugh M.E.. The Legume Information System (LIS): An integrated information resource for comparative legume biology. Nucleic Acids Res. 2005; 33:D660–D665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Proost S., Van Bel M., Vaneechoutte D., Van De Peer Y., Inzé D., Mueller-Roeber B., Vandepoele K.. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 2015; 43:D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al. Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schuler G.D. Sequence mapping by electronic PCR. Genome Res. 1997; 7:541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bustin S.a., Benes V., Garson J.a., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 27. Mungall C.J., Emmert D.B., Gelbart W.M., de Grey A., Letovsky S., Lewis S.E., Rubin G.M., Shu S.Q., Wiel C., Zhang P. et al. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007; 23:i337–i346. [DOI] [PubMed] [Google Scholar]
- 28. Sanderson L.A., Ficklin S.P., Cheng C.H., Jung S., Feltus F.A., Bett K.E., Main D.. Tripal v1.1: a standards-based toolkit for construction of online genetic and genomic databases. Database (Oxford). 2013; 2013:bat075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li T., Pan G.-Q., Vossbrinck C.R., Xu J.-S., Li C.-F., Chen J., Long M.-X., Yang M., Xu X.-F., Xu C. et al. SilkPathDB: a comprehensive resource for the study of silkworm pathogens. Database (Oxford). 2017; 2017:bax001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.