Abstract
Extensive efforts have been directed at the discovery, investigation and clinical monitoring of targeted therapeutics. These efforts may be facilitated by the convenient access of the genetic, proteomic, interactive and other aspects of the therapeutic targets. Here, we describe an update of the Therapeutic target database (TTD) previously featured in NAR. This update includes: (i) 2000 drug resistance mutations in 83 targets and 104 target/drug regulatory genes, which are resistant to 228 drugs targeting 63 diseases (49 targets of 61 drugs with patient prevalence data); (ii) differential expression profiles of 758 targets in the disease-relevant drug-targeted tissue of 12 615 patients of 70 diseases; (iii) expression profiles of 629 targets in the non-targeted tissues of 2565 healthy individuals; (iv) 1008 target combinations of 1764 drugs and the 1604 target combination of 664 multi-target drugs; (v) additional 48 successful, 398 clinical trial and 21 research targets, 473 approved, 812 clinical trial and 1120 experimental drugs, and (vi) ICD-10-CM and ICD-9-CM codes for additional 482 targets and 262 drugs against 98 disease conditions. This update makes TTD more useful for facilitating the patient focused research, discovery and clinical investigations of the targeted therapeutics. TTD is accessible at http://bidd.nus.edu.sg/group/ttd/ttd.asp.
INTRODUCTION
Collective efforts in the development and practice of modern medicines by the research communities, pharmaceutical companies and clinical communities have been primarily directed at the discovery, investigation and clinical monitoring of the targeted therapeutics (1–6). These efforts have been partly facilitated by the knowledge of the genomics, transcriptomics, structures, interactions, biological systems and functional profiles of the therapeutic targets (7–10). In particular, a number of freely accessible databases complementarily provide comprehensive information about the therapeutic targets with additional information about the clinical trial drugs (11–13), biomarkers (11), molecular activity data (11,13–15), drug-binding sites (16,17) and target-affiliated biological pathways (11,18) of the targets, as well as the therapeutics (12,14,15,19,20) and ADME-Tox properties (12) of the targeted drugs.
The clinical efficacies of the targeted therapeutics and the outcomes of target evaluation studies frequently depend on the states of drug-resistance mutations (21,22) and the gene expression profiles (23,24) of the targets and their regulators in the patients. Moreover, the multi-target drugs (25,26) and drug combinations (25,27,28) directed at specific target combinations have been extensively tested and used for enhanced therapeutics. The knowledge of the drug resistance mutations (21,29), patient gene expression profiles (30) and the target combinations of the multi-target drugs (26,31) and drug combinations (27) has facilitated our understanding of the mechanisms of drug therapeutics, the assessment of druggability of the targets (7,32,33) and the development and practices of stratified and precision medicines (34–36). However, the relevant information is insufficiently covered by the existing database. As of August 2017, the Cancer Genome Interpreter (https://www.cancergenomeinterpreter.org/), Catalogue of Somatic Mutations in Cancer (37), Comprehensive Antibiotic Resistance Database (38), HIV Drug Resistance Mutation Database (39) and Tuberculosis Drug Resistance Mutation Database (40) collectively provide 1688 drug resistance mutations in 85 targets of 90 drugs, which are significantly less than the reported drug resistance mutation studies for ∼200 targets and ∼200 drugs we found from the literature. While the GEO (41) and TCGA (42) databases provide comprehensive genome-scale gene expression data of the patients of different diseases and healthy individuals, the therapeutic targets and regulators are not explicitly marked for convenient access of the relevant data. The DCDB database (43) contains the 761 target combinations of 1363 drug combinations, which are significantly <1008 target combinations of 2042 drug combinations we searched from literature and the ClinicalTrials.gov service (https://clinicaltrials.gov/).
To provide more comprehensive information about the drug resistance mutations, gene expressions and target combinations data for the targets and drugs, we updated the Therapeutic target database (TTD, http://bidd.nus.edu.sg/group/ttd/ttd.asp) with several major improvements. The first improvement is the inclusion of the literature-reported drug resistance mutations in 83 targets (40 successful, 34 clinical trial and 9 research targets) and 104 target/drug regulatory genes, which reportedly are resistant to 228 drugs (128 approved, 56 clinical trial and 44 investigative) targeting 63 diseases. While available from the literature search, the prevalence of the drug resistance mutations in the targeted patient populations was also included. The second is the inclusion of the differential expression profiles of 309 successful and 449 clinical trial targets in the disease-relevant drug targeted tissue of 12 615 patients of 70 diseases. The differential expression profile contains the gene expression levels of the targeted individual patients and those of the healthy individuals. The third is the inclusion of the expression profiles of 372 successful and 257 clinical trial targets in the non-targeted tissues of 2565 healthy individuals. Typically, for each target, the average gene expression levels in 37–45 tissues are provided. The fourth is the inclusion of 1008 target combinations of 626 and 1138 combinations of approved and clinical trial drugs, and the 1604 target combination of 122 approved, 542 clinical trial and 25 333 research multi-target drugs. Moreover, we also added additional 48 successful, 398 clinical trial and 21 research targets, 473 approved, 812 clinical trial and 1120 experimental drugs, and the ICD-10-CM and ICD-9-CM codes for additional 482 targets and 262 drugs against 98 disease conditions. Table 1 provides the statistics of the latest TTD target and drug data with respect to that of previous release.
Table 1. Statistics of the target and drug data in TTD.
| 2018 update | 2016 update | ||
|---|---|---|---|
| Statistic of targets | Number of all targets | 3101 | 2589 |
| Number of successful targets | 445 | 397 | |
| Number of clinical trial targets | 1121 | 723 | |
| Number of research targets | 1535 | 1469 | |
| Statistic of drugs | Number of all drugs | 34 019 | 31 614 |
| Number of approved drugs | 2544 | 2071 | |
| Number of clinical trial drugs | 8103 | 7291 | |
| Number of investigative drugs | 18 923 | 17 803 | |
| Number of bi-specific antibody | 21 | 0 | |
| Number of stem cell drugs | 10 | 0 | |
| Number of multi-target agents | 26 459 | 26 368 | |
| Number of drugs withdrawn from the market | 158 | 154 | |
| Number of drugs discontinued in clinical trial | 2349 | 2237 | |
| Number of pre-clinical drugs | 417 | 357 | |
| Number of drugs terminated in unspecified investigative stage | 1929 | 1701 | |
| Number of small molecular drugs with available structure | 21 936 | 17 356 | |
| Number of approved drugs with available structure | 2326 | 1896 | |
| Number of clinical trial drugs with available structure | 4258 | 2161 | |
| Number of investigative with available structure | 15 352 | 13 308 |
DRUG RESISTANCE MUTATIONS
Drug resistance is a global public health problem particularly for the treatment of infectious diseases and cancers (21). The clinical efficacy of the antibiotic drugs is rapidly decreasing partly due to the occurrence of drug resistance mutations in the targets or the target/drug regulatory genes in the bacteria (44). Cancers’ acquisition of drug resistance is partly linked to specific somatic mutations of the anticancer targets and the target/drug regulatory genes (45). The knowledge of the drug resistance mutations in the targets and target/drug regulatory genes are important for the understanding of the mechanisms of drug therapeutic efficacies (21,45), the prediction of drug resistance mutations (29) and the development and practices of stratified and precision medicines (34).
The drug resistance mutations were searched from the PubMed database using the combinations of the keywords ‘resistance’, ‘resistant’, ‘mutation’, ‘mutations’ and the name and synonyms of each of the targets and drugs in the TTD database (11). The identified literatures were manually evaluated to extract the relevant drug resistance mutations in each target or the target/drug regulatory genes. The collected information includes the drug resistance mutations and mutation types (missense mutation, nonsense mutation, deletion, frameshift and insertion), level of resistance (defined by the fold-change of the IC50 values with and without the respective mutation) and the prevalence of mutations in the patients of the drug targeted diseases. The wild-type sequence of each target or target/drug regulatory gene product from the UniProt database (46) was also included for reference. Overall, there are 2000 drug resistance mutations in 83 targets (40 successful, 34 clinical trial and 9 research) and 104 target/drug regulatory genes, which are resistant to 228 drugs (128 approved, 56 clinical trial and 44 investigative) targeting 63 diseases (49 targets of 61 drugs with patient prevalence data).
TARGET GENE EXPRESSION
Drug interaction with its target frequently induces a positive or negative feedback loop that adjusts the expression level of the target (30). Systematic gene expression analysis of the successful and phase III clinical trial targets across a diverse collection of normal human tissues have shown that most targets are expressed in a disease-affected tissue under healthy conditions (23). The human tissue distribution profile of a target, determined by the level of the target expression in different human tissues, is a key druggability indicator (7). This indicator together with other druggability indicators have been used to prospectively predict 16 promising targets among 31 phase III targets of small molecule drugs in 2009, 10 of which have become successful following the publication of the target assessments (32). Therefore, there is a need for the gene expression data of the targets in both the disease-relevant drug targeted tissue and other tissues of the patients and healthy individuals.
The relevant gene expression data for the targets were searched by the following procedures. First, 2538 series records of human gene expression raw data based on the Affymetrix U133 Plus 2.0 platform were collected from the GEO database (41), and their corresponding disease and tissue information were recorded from the sample annotation. Then, 569 series records of 70 diseases and 45 tissues covered by TTD (11) were selected. Those samples from the same disease and the same tissue were selected for meta-analysis of the gene expression profiles. Second, data pre-processing and quality control were conducted based on the normalized unscaled standard error and the relative log expression (47,48), which retained 34 355 samples from all gene expression raw data. Thirdly, those gene expression data from the same disease and the same tissue were combined and normalized using the robust multiarray average, quantile normalization, perfect match correction and median polish, based on the R package affy (49). Fourth, fold-change and Z-score transformation allowed the comparison of data across a wide range of experiments, by choosing a baseline of array expression that had the median intensities (excluded the highest and lowest 2% of probe intensities), and then standardizing all arrays to this baseline (50). In this study, fold change, Z-scores and Student’s t-test were applied to reveal the differential expression of targets among three sample groups. The first group covers target gene expression profiles of the patients in the disease section of the disease-relevant drug targeted tissue, the second group covers the profiles of the patients in the normal section of the tissue adjacent to the disease section and the third group covers the profiles in the tissue of healthy individuals (51). Finally, the R package ggplot2 was further applied to generate the gene expression plots which were displayed on the TTD official website.
TARGET COMBINATIONS OF MULTI-TARGET DRUGS AND DRUG COMBINATIONS
Targeted therapeutics directed at individual target are frequently insufficient against heterogenetic diseases like cancers, or diseases that affect multiple tissues or cell types such as diabetes and immune-inflammatory disorders. Multi-target drugs (25,26,31) and drug combinations (27,28,52) have been increasingly tested and used for the improved treatment of these heterogenetic and multi-tissue/multi-cell-type diseases, and for overcoming drug resistances. Knowledge of the multiple targets of multi-target drugs are important for determining the clinical therapeutic efficacy particularly in the patients of active drug-bypass genes (26), and for network analysis of the therapeutic efficiency of the multi-targeting actions (31). Knowledge of the multiple targets of the drug combinations is important for optimization of drug combinations (52), selection of different mechanisms of action to minimize resistance (28) and understanding of the mechanisms of the synergistic drug combinations (27). Hence, a resource with convenient access of the target combinations of the multi-target drugs and drug combinations is useful for facilitating the relevant investigations.
The target combinations of the multi-target drugs and drug combinations were collected from the existing database and literature. Specifically, multi-target drugs were from the TTD (11) and ClinicalTrials.gov databases, followed by the literature search for finding their primary therapeutic efficacy targets, and the developmental status of these drugs were also collected. Drug combinations were obtained from the Drugs@FDA (https://www.accessdata.fda.gov/scripts/cder/daf/), ClinicalTrials.gov service and additional literature search of PubMed (53) using the combinations of keywords ‘drug’, ‘combination’, ‘combine’, ‘plus’ and individual target or drug name or synonyms. The primary therapeutic target of each drug in a specific drug combination was identified based on TTD data. By combining the primary therapeutic targets of all drugs in a certain drug combination, a target combination was generated. The biochemical class, structural fold and pathway information of each target in a specific target combination were further searched from the UniProt database (46), CATH Gene3D (54) and KEGG database (55) respectively. By using these information, target combinations can be classified and searched according to their target family pairs, target structural fold pairs and pathway pairs, and such search options are provided in this update of TTD to facilitate the customized search of each target combination.
THE NEW TTD DATA STORAGE AND ACCESS FACILITIES
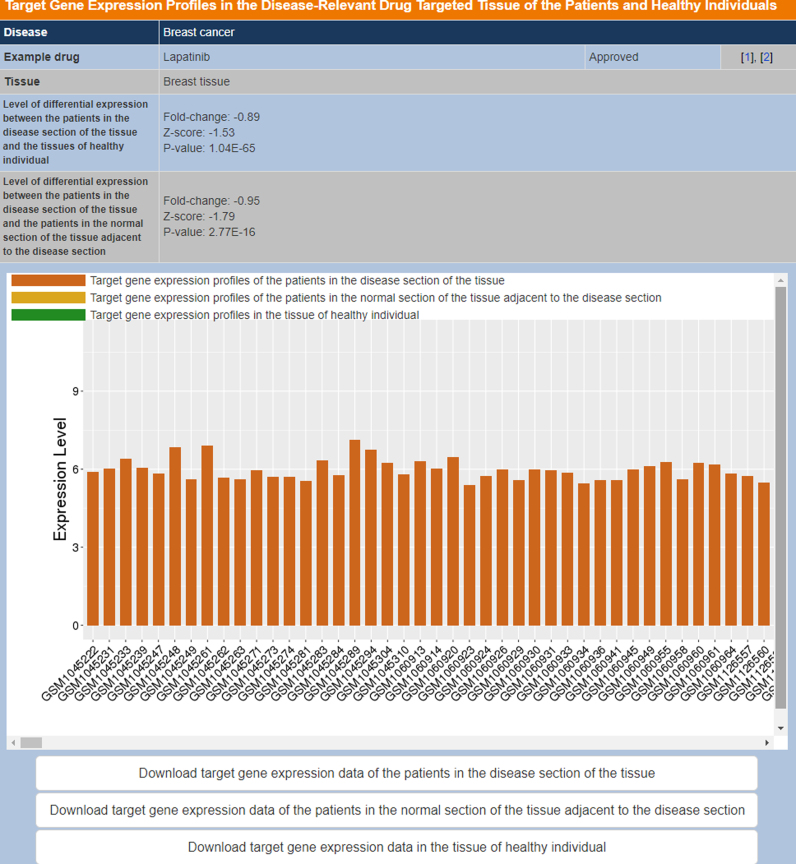
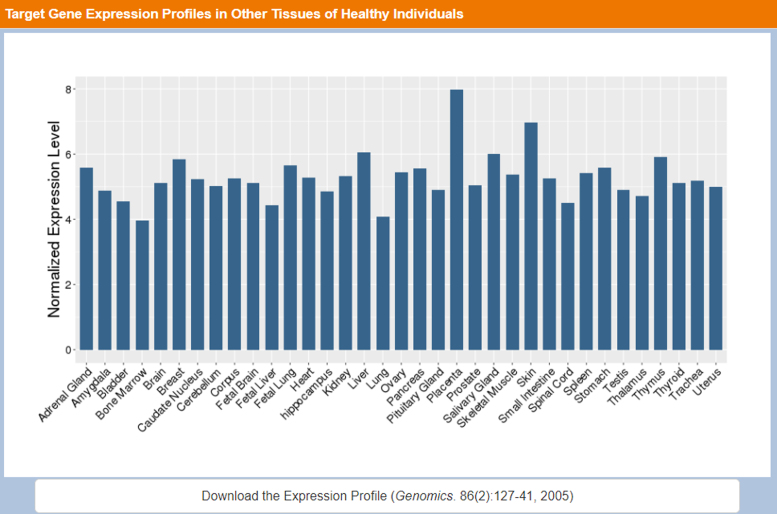
The TTD database architecture and interface were redesigned for facilitating more convenient access of the new as well as existing data. In particular, we employed Drupal as the database platform for enhanced data storage and extraction. The more accelerated data access and transmission was made possible by using the cloud platform of Aliyun located in the Silicon Valley of USA. In the new TTD interface, the newly added drug resistance mutation and target expression data can be accessed through the Patient Data manual bar, and the target combination information can be accessed through the Targets/Drugs Group manual bar. The Drugs Group manual bar also includes multi-target agents and nature-derived drugs search option. The Advanced Search manual bar includes customized search, target similarity search, drug similarity search and pathway search options. The drug similarity search facility is further divided into small molecule drug structural similarity search, anti-sense drug nucleotide sequence similarity search and antibody drug amino acid sequence similarity search based on molecular fingerprint Tanimoto coefficient (11), blastn (56) and blastp (56) similarity search methods respectively. Moreover, the JSME Molecule Editor (57) were added for facilitating users to draw a molecule and subsequently search the TTD drug entries that are similar in structure to the input molecule. The Model & Study Data manual bar includes target validation data and QSAR models search options. Figure 1 shows the section of the search result page that presents the gene expression profiles in the disease-relevant drug-targeted tissue of the patients and healthy individuals. Figure 2 shows the section of the search result page that presents the gene expression profiles in the other tissues of healthy individuals. Figure 3 shows the search result page of drug resistance mutations.
Figure 1.
The section of the search result page that presents the gene expression profiles in the disease-relevant drug-targeted tissue of the patients and healthy individuals.
Figure 2.
The section of the search result page that presents the gene expression profiles in the other tissues of healthy individuals.
Figure 3.
The search result page of drug resistance mutations.
PERSPECTIVES
The continuous development of the relevant databases (11,24,58–60) has provided richer and increasingly useful information resources for the therapeutic targets and drugs to better serve the drug discovery and clinical medicine communities. With increasing movement of the modern therapeutics toward stratified and personalized medicines (34), extensive efforts from the research, industry, clinical, regulatory and management communities and the chemistry, biology, pharmaceutics and medicine disciplines have been collectively directed at the discovery, investigation, application, monitoring and management of targeted therapeutics (1). and the novel treatment strategies such as multi-target drugs (25,26,31), drug combinations (27,28,52), monoclonal antibodies (61) and RNA therapeutics (62) for enhanced therapeutics and for overcoming drug resistances (21,22). The TTD and other databases may be further expanded to incorporate the newly derived data and novel knowledge to cater the new needs for the development of novel therapeutics.
FUNDING
Singapore Academic Research Fund [R-148–000-208–112, R-148–000-230–114, R-148–000-239–114]; Precision Medicine Project of the National Key Research and Development Plan of China [2016YFC0902200]; Innovation Project on Industrial Generic Key Technologies of Chongqing [cstc2015zdcy-ztzx120003]. Funding for open access charge: Precision Medicine Project of the National Key Research and Development Plan of China [2016YFC0902200]; Innovation Project on Industrial Generic Key Technologies of Chongqing [cstc2015zdcy-ztzx120003].
Conflict of interest statement. None declared.
REFERENCES
- 1. Munos B. Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov. 2009; 8:959–968. [DOI] [PubMed] [Google Scholar]
- 2. Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G., Karlsson A., Al-Lazikani B., Hersey A., Oprea T.I. et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017; 16:19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Imming P., Sinning C., Meyer A.. Drugs, their targets and the nature and number of drug targets. Nat. Rev. Drug Discov. 2006; 5:821–834. [DOI] [PubMed] [Google Scholar]
- 4. Zhang X.D., Song J., Bork P., Zhao X.M.. The exploration of network motifs as potential drug targets from post-translational regulatory networks. Sci. Rep. 2016; 6:20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee A., Lee K., Kim D.. Using reverse docking for target identification and its applications for drug discovery. Expert Opin. Drug Discov. 2016; 11:707–715. [DOI] [PubMed] [Google Scholar]
- 6. Wang P., Zhang X., Fu T., Li S., Li B., Xue W., Yao X., Chen Y., Zhu F.. Differentiating physicochemical properties between addictive and nonaddictive ADHD drugs revealed by molecular dynamics simulation studies. ACS Chem. Neurosci. 2017; 8:1416–1428. [DOI] [PubMed] [Google Scholar]
- 7. Zheng C.J., Han L.Y., Yap C.W., Ji Z.L., Cao Z.W., Chen Y.Z.. Therapeutic targets: progress of their exploration and investigation of their characteristics. Pharmacol. Rev. 2006; 58:259–279. [DOI] [PubMed] [Google Scholar]
- 8. Rask-Andersen M., Almen M.S., Schioth H.B.. Trends in the exploitation of novel drug targets. Nat. Rev. Drug Discov. 2011; 10:579–590. [DOI] [PubMed] [Google Scholar]
- 9. Chen B., Butte A.J.. Leveraging big data to transform target selection and drug discovery. Clin. Pharmacol. Ther. 2016; 99:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang P., Fu T., Zhang X., Yang F., Zheng G., Xue W., Chen Y., Yao X., Zhu F.. Differentiating physicochemical properties between NDRIs and sNRIs clinically important for the treatment of ADHD. Biochim. Biophys. Acta. 2017; 1861:2766–2777. [DOI] [PubMed] [Google Scholar]
- 11. Yang H., Qin C., Li Y.H., Tao L., Zhou J., Yu C.Y., Xu F., Chen Z., Zhu F., Chen Y.Z.. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016; 44:D1069–D1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014; 42:D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibrian-Uhalte E. et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017; 45:D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Southan C., Sharman J.L., Benson H.E., Faccenda E., Pawson A.J., Alexander S.P., Buneman O.P., Davenport A.P., McGrath J.C., Peters J.A. et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res. 2016; 44:D1054–D1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ursu O., Holmes J., Knockel J., Bologa C.G., Yang J.J., Mathias S.L., Nelson S.J., Oprea T.I.. DrugCentral: online drug compendium. Nucleic Acids Res. 2017; 45:D932–D939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ito J., Ikeda K., Yamada K., Mizuguchi K., Tomii K.. PoSSuM v.2.0: data update and a new function for investigating ligand analogs and target proteins of small-molecule drugs. Nucleic Acids Res. 2015; 43:D392–D398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang C., Hu G., Wang K., Brylinski M., Xie L., Kurgan L.. PDID: database of molecular-level putative protein-drug interactions in the structural human proteome. Bioinformatics. 2016; 32:579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim P., Cheng F., Zhao J., Zhao Z.. ccmGDB: a database for cancer cell metabolism genes. Nucleic Acids Res. 2016; 44:D959–D968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gohlke B.O., Nickel J., Otto R., Dunkel M., Preissner R.. CancerResource–updated database of cancer-relevant proteins, mutations and interacting drugs. Nucleic Acids Res. 2016; 44:D932–D937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu F., Qin C., Tao L., Liu X., Shi Z., Ma X., Jia J., Tan Y., Cui C., Lin J. et al. Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12943–12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hughes D., Andersson D.I.. Evolutionary consequences of drug resistance: shared principles across diverse targets and organisms. Nat. Rev. Genet. 2015; 16:459–471. [DOI] [PubMed] [Google Scholar]
- 22. Schmitt M.W., Loeb L.A., Salk J.J.. The influence of subclonal resistance mutations on targeted cancer therapy. Nat Rev. Clin Oncol. 2016; 13:335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar V., Sanseau P., Simola D.F., Hurle M.R., Agarwal P.. Systematic analysis of drug targets confirms expression in disease-relevant tissues. Sci. Rep. 2016; 6:36205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu F., Han B., Kumar P., Liu X., Ma X., Wei X., Huang L., Guo Y., Han L., Zheng C. et al. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 2010; 38:D787–D791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zimmermann G.R., Lehar J., Keith C.T.. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov. Today. 2007; 12:34–42. [DOI] [PubMed] [Google Scholar]
- 26. Tao L., Zhu F., Xu F., Chen Z., Jiang Y.Y., Chen Y.Z.. Co-targeting cancer drug escape pathways confers clinical advantage for multi-target anticancer drugs. Pharmacol. Res. 2015; 102:123–131. [DOI] [PubMed] [Google Scholar]
- 27. Jia J., Zhu F., Ma X., Cao Z., Cao Z.W., Li Y., Li Y.X., Chen Y.Z.. Mechanisms of drug combinations: interaction and network perspectives. Nat. Rev. Drug Discov. 2009; 8:111–128. [DOI] [PubMed] [Google Scholar]
- 28. Kummar S., Chen H.X., Wright J., Holbeck S., Millin M.D., Tomaszewski J., Zweibel J., Collins J., Doroshow J.H.. Utilizing targeted cancer therapeutic agents in combination: novel approaches and urgent requirements. Nat. Rev. Drug Discov. 2010; 9:843–856. [DOI] [PubMed] [Google Scholar]
- 29. Cao Z.W., Han L.Y., Zheng C.J., Ji Z.L., Chen X., Lin H.H., Chen Y.Z.. Computer prediction of drug resistance mutations in proteins. Drug Discov. Today. 2005; 10:521–529. [DOI] [PubMed] [Google Scholar]
- 30. Iskar M., Campillos M., Kuhn M., Jensen L.J., van Noort V., Bork P.. Drug-induced regulation of target expression. PLoS Comput. Biol. 2010; 6:e1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Csermely P., Agoston V., Pongor S.. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol. Sci. 2005; 26:178–182. [DOI] [PubMed] [Google Scholar]
- 32. Zhu F., Han L., Zheng C., Xie B., Tammi M.T., Yang S., Wei Y., Chen Y.. What are next generation innovative therapeutic targets? clues from genetic, structural, physicochemical, and systems profiles of successful targets. J. Pharmacol. Exp. Ther. 2009; 330:304–315. [DOI] [PubMed] [Google Scholar]
- 33. Xue W., Wang P., Li B., Li Y., Xu X., Yang F., Yao X., Chen Y.Z., Xu F., Zhu F.. Identification of the inhibitory mechanism of FDA approved selective serotonin reuptake inhibitors: an insight from molecular dynamics simulation study. Phys. Chem. Chem. Phys. 2016; 18:3260–3271. [DOI] [PubMed] [Google Scholar]
- 34. Hauschild A., Grob J.J., Demidov L.V., Jouary T., Gutzmer R., Millward M., Rutkowski P., Blank C.U., Miller W.H. Jr, Kaempgen E. et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012; 380:358–365. [DOI] [PubMed] [Google Scholar]
- 35. Li B., Tang J., Yang Q., Li S., Cui X., Li Y., Chen Y., Xue W., Li X., Zhu F.. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017; 45:W162–W170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li B., Tang J., Yang Q., Cui X., Li S., Chen S., Cao Q., Xue W., Chen N., Zhu F.. Performance evaluation and online realization of data-driven normalization methods used in LC/MS based untargeted metabolomics analysis. Sci. Rep. 2016; 6:38881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017; 45:D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., Lago B.A., Dave B.M., Pereira S., Sharma A.N. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017; 45:D566–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang M.W., Liu T.F., Shafer R.W.. The HIVdb system for HIV-1 genotypic resistance interpretation. Intervirology. 2012; 55:98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sandgren A., Strong M., Muthukrishnan P., Weiner B.K., Church G.M., Murray M.B.. Tuberculosis drug resistance mutation database. PLoS Med. 2009; 6:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Y., Hu B., Fu C., Chen X.. DCDB: drug combination database. Bioinformatics. 2010; 26:587–588. [DOI] [PubMed] [Google Scholar]
- 44. MacLean R.C., Hall A.R., Perron G.G., Buckling A.. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat. Rev. Genet. 2010; 11:405–414. [DOI] [PubMed] [Google Scholar]
- 45. Bell D.W., Gore I., Okimoto R.A., Godin-Heymann N., Sordella R., Mulloy R., Sharma S.V., Brannigan B.W., Mohapatra G., Settleman J. et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat. Genet. 2005; 37:1315–1316. [DOI] [PubMed] [Google Scholar]
- 46. UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lukk M., Kapushesky M., Nikkila J., Parkinson H., Goncalves A., Huber W., Ukkonen E., Brazma A.. A global map of human gene expression. Nat. Biotechnol. 2010; 28:322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Torrente A., Lukk M., Xue V., Parkinson H., Rung J., Brazma A.. Identification of cancer related genes using a comprehensive map of human gene expression. PLoS One. 2016; 11:e0157484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gautier L., Cope L., Bolstad B.M., Irizarry R.A.. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004; 20:307–315. [DOI] [PubMed] [Google Scholar]
- 50. Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P.. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003; 19:185–193. [DOI] [PubMed] [Google Scholar]
- 51. Cheadle C., Vawter M.P., Freed W.J., Becker K.G.. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 2003; 5:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dancey J.E., Chen H.X.. Strategies for optimizing combinations of molecularly targeted anticancer agents. Nat. Rev. Drug Discov. 2006; 5:649–659. [DOI] [PubMed] [Google Scholar]
- 53. Coordinators N.R. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2017; 45:D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sillitoe I., Lewis T.E., Cuff A., Das S., Ashford P., Dawson N.L., Furnham N., Laskowski R.A., Lee D., Lees J.G. et al. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 2015; 43:D376–D381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M.. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016; 44:D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 57. Bienfait B., Ertl P.. JSME: a free molecule editor in JavaScript. J. Cheminform. 2013; 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qin C., Zhang C., Zhu F., Xu F., Chen S.Y., Zhang P., Li Y.H., Yang S.Y., Wei Y.Q., Tao L. et al. Therapeutic target database update 2014: a resource for targeted therapeutics. Nucleic Acids Res. 2014; 42:D1118–D1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen X., Ji Z.L., Chen Y.Z.. TTD: therapeutic target database. Nucleic Acids Res. 2002; 30:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhu F., Shi Z., Qin C., Tao L., Liu X., Xu F., Zhang L., Song Y., Liu X., Zhang J. et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012; 40:D1128–D1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Weiner G.J. Building better monoclonal antibody-based therapeutics. Nat. Rev. Cancer. 2015; 15:361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rupaimoole R., Slack F.J.. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017; 16:203–222. [DOI] [PubMed] [Google Scholar]