Abstract
The Plant Resistance Genes database (PRGdb; http://prgdb.org) has been redesigned with a new user interface, new sections, new tools and new data for genetic improvement, allowing easy access not only to the plant science research community but also to breeders who want to improve plant disease resistance. The home page offers an overview of easy-to-read search boxes that streamline data queries and directly show plant species for which data from candidate or cloned genes have been collected. Bulk data files and curated resistance gene annotations are made available for each plant species hosted. The new Gene Model view offers detailed information on each cloned resistance gene structure to highlight shared attributes with other genes. PRGdb 3.0 offers 153 reference resistance genes and 177 072 annotated candidate Pathogen Receptor Genes (PRGs). Compared to the previous release, the number of putative genes has been increased from 106 to 177 K from 76 sequenced Viridiplantae and algae genomes. The DRAGO 2 tool, which automatically annotates and predicts (PRGs) from DNA and amino acid with high accuracy and sensitivity, has been added. BLAST search has been implemented to offer users the opportunity to annotate and compare their own sequences. The improved section on plant diseases displays useful information linked to genes and genomes to connect complementary data and better address specific needs. Through, a revised and enlarged collection of data, the development of new tools and a renewed portal, PRGdb 3.0 engages the plant science community in developing a consensus plan to improve knowledge and strategies to fight diseases that afflict main crops and other plants.
INTRODUCTION
Plant crops are susceptible to a large number of pathogens, including bacteria, fungi, oomycetes, viruses, nematodes and insects. In the last 70 years, breeding efforts provided a continuous supply of cultivars with improved yield and quality traits (1), though damage brought by plant pests and diseases notably reduces the global crop yield (2). Moreover, climate changes have facilitated the movement of numerous organisms and in turn have triggered new diseases that could develop into uncontrollable epidemics and jeopardize food security (3). For these reasons, plant breeders and researchers are highly committed to searching for plant disease resistance mechanisms. Plants possess a sophisticated immune system based on their ability to recognize phytopathogens. The activation of this system is based on the presence of specific receptors encoded by the so-called pathogen recognition genes (PRGs). The proteins encoded by the PRGs share common domains such as coiled-coil (CC), nucleotide binding region (NB), Toll-interleukin region (TIR), leucine rich region (LRR) and kinase domain (K). The cytoplasmic NB-LRR genes are divided into two classes: TNL (TIR-NB-LRR) and CNL (CC-NB-LRR) which possess, either the TIR or the CC domains respectively. Transmembrane receptor proteins containing kinase and LRR domains, such as receptor-like proteins (RLP) and the receptor-like kinases (RLK), are also involved (4). Omics and bioinformatics resources have greatly enhanced the identification, the genetic selection and the cloning of novel PRGs in the last few years. New approaches for exploring resistance genes datasets proved useful for shedding light on their molecular and evolutionary mechanisms and for facilitating the design of diagnostic tests, comparative analyses and new breeding programs (5). According to Food and Agriculture Organization (FAO) guidelines, omics data will be extremely important to develop more efficient plant cultivars, which are necessary for a new ‘greener revolution’ (6).
In the last years, several online omics platforms have been offered to facilitate the exploration and use of plant resistance genes. These platforms offer information related to specific organisms (7,8) or to investigate particular plant traits (9), others offer omics data and integrated comparative tools (10). In this context, the Pathogen Recognition Genes database (PRGdb) represents an important reference site and repository for people working in the plant biotic resistance field and in the last year it improved understanding and findings in this research area. Since 2009, PRGdb supported researchers and plant breeders in detecting new plant disease resistance sources useful for crop improvement.
In this work, we present version 3.0 of PRGdb (http://prgdb.org), an update that contains new cloned and candidate pathogen recognition genes, allowing users to browse the manual annotated PR genes, PR gene families and avirulence (Avr) genes. A PRG annotation conducted on 76 Viridiplantae and algae proteomes is also provided. Furthermore, we offer users a new, automatic system to annotate and predict PRGs from DNA and amino acid (AA) sequences with a significant improvement in accuracy and sensitivity in comparison to our previous tool. These and other features are available in the new PRGdb user-friendly website.
MATERIALS AND METHODS
New cloned resistance genes, Viridiplantae and algae proteomes
An extensive bibliographic search was performed to retrieve the new cloned resistance genes from 1 January 2014 (last update of PRGdb 2.0 (11)) to 1 August 2016. In addition, public databases such as Phytozome (12), Ensembl Plants (13) and NCBI genome db (14) were explored to upload the most recently released Viridiplantae and algae proteomes, retrieving a total of 76 proteome sequences (see the list of these proteomes in Supplementary Table S1).
Construction of hidden Markov models (HMMs) for R genes
The AA sequences of the cloned PRGs were incorporated in four FASTA files according to the PRG class they belong to (CNLs, TNLs, RLPs and RLKs). Afterward, a multiple sequence alignment (MSA) for each PRG class FASTA was performed using MUSCLE 3.6 (15). Then, these MSAs were used as a base for the creation of hidden Markov models (HMM) using the HMMER v3 package (16). An in-house PERL script was used to filter the best alignments and create the HMM modules using hmmbuild command. Alignments in a given position with a minimum BLOSUM62 score of +1 in the AA comparison and a minimum of 10 AAs in length were considered. Thus, a total of 60 HMM modules were built. A double check was performed using hmmsearch with the initial FASTA files as input against these HMM modules. To further classify these HMM modules as domains of resistance classes, each domain sequence from a certain class was extracted as a FASTA file from the above-mentioned MSA using Geneious R9 desktop software (17). Then, each of these FASTA files were employed as inputs to hmmsearch in order to label the HMM modules according to the domains they target. Finally, the HMM modules that did not have a match were further tested with jackhmmer tool (15) to find matches in Uniprot database (18).
Pathogen recognition gene analysis and gene orthology (DRAGO 2) pipeline
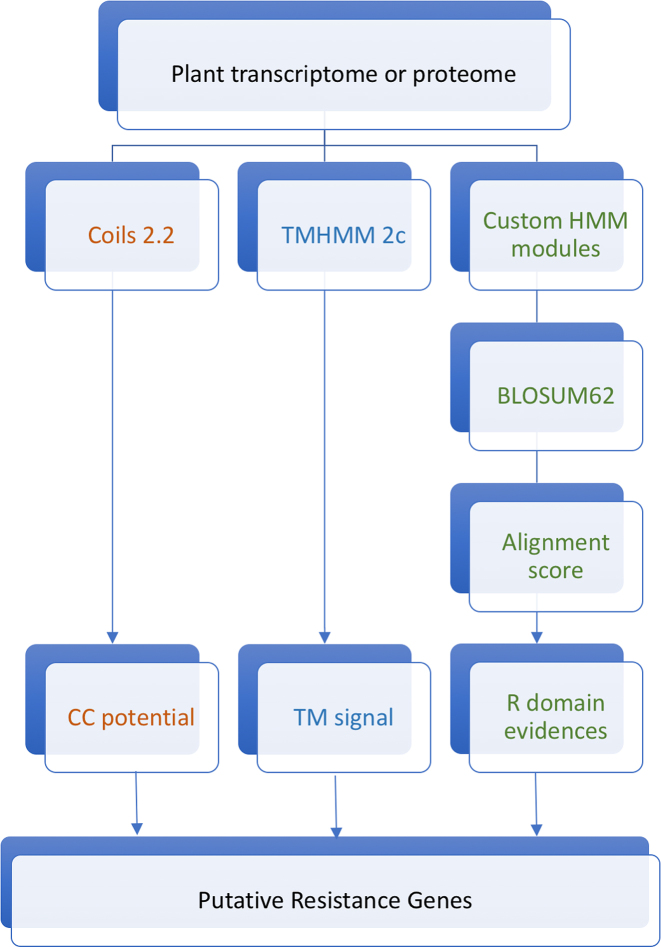
The core of pathogen recognition genes analysis and gene orthology (DRAGO 2) pipeline consists of a PERL script that predicts putative PRGs within a plant transcriptome/proteome FASTA file (Figure 1). In a first round, DRAGO 2 was executed with the cloned PRG FASTA file as an input to define the normalization value and the minimum score thresholds. Specifically, the previously created 60 HMM modules are used by DRAGO 2 to detect LRR, Kinase, NBS and TIR domains and compute the alignment score of the different hits based on a BLOSUM62 matrix. The normalization value is the absolute smallest similarity score found among the input sequences considering all domains. The minimum score thresholds are calculated from the smallest similarity score reported in a specific domain among the input sequences. Once these values are known, DRAGO 2 can be launched on any transcriptome or proteome. Apart from detecting the mentioned domains, DRAGO 2 is also able to detect CC domains and TM domains using COILS 2.2 program (19) and TMHMM 2.0c program (20). DRAGO 2 generates two output files: a numeric matrix that represents the similarity score of every single protein input to each HMM profile, and a JSON or TSB format file with the domain name, start position, end position, resistance class and identification for every putative plant resistance protein.
Figure 1.
DRAGO 2 pipeline: an overview of tools and script that predicts putative resistance genes within a plant transcriptome/proteome FASTA file.
DRAGO 2 validation
To validate this tool, DRAGO 2 was executed over the proteome of the well-studied Arabidopsis thaliana organism to predict plant pathogen recognition proteins. The FASTA file of these putative PR sequences was further analyzed by InterProScan program version 5.16 (21) with the aim of comparing its results with those from our tool. InterProScan was called using PfamA-26.0 and Coils-2.2, with the other parameters set to default.
Relational database
The PRG data was imported into a MySQL (5.5) based relational database hosted in an Ubuntu server (14.04). The web application was developed using NodeJS technology with the ExpressJS web development framework in the back-end and HTML5, CSS3, JavaScript technologies in the front-end, besides importing libraries and frameworks such as Bootstrap or JQuery. PRGdb 3.0 is freely accessible through a web interface at the following address: http://www.prgdb.org.
RESULTS
Overview of improved web portal display and features
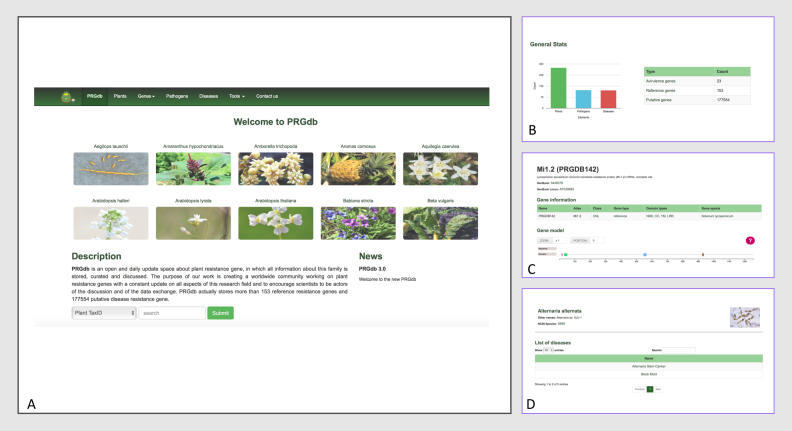
One of the major goals of this update was the creation of a more interactive website for the plant disease resistance community. We have modernized the PRGdb interface to improve user experience, making contents more accessible and greatly expanding the amount of pages and sections. We have re-engineered the home page to summarize the different types of information gathered in the portal (Figure 2A and B). Users can navigate through the site using the dropdown menu, showing from left to right: PRGdb home, species, genes (which includes Reference PRGs, Putative PRGs, Avr genes and All resistance genes), Pathogens, Diseases, tools (which includes DRAGO 2 and BLAST) and Contact us. In the middle of the home page, plants having PRGs are displayed and associated with a picture. In the plant images section, clicking on a specific photo expands the details on a given plant. In the general stats section, users will find a chart and a table to quickly visualize the amount of information stored in our database. The number of plants (green bar), pathogens (blue bar) and diseases (red bar) stored in our database are shown in a chart, whereas number of Avr genes, reference genes and putative genes are shown in a table. Finally, the last section displays news related to the web application and plant disease resistance.
Figure 2.
An overview of the PRGdb version 3.0. (A) Home page and Search tool; (B) General stats; (C) Gene page with interactive gene model system and related gene info; (D) example of Pathogens page with disease information.
Plant, pathogen and gene model page
The ‘Plants’ page shows a list of all the organisms stored in this database. This list can be dynamically consulted searching by plant name. In each plant species page users can find statistics on the number of reference and putative pathogen recognition genes for each class in that species, as well as other information such as its NCBI accession number or a photo.
The ‘Genes’ page shows a summary of all the information attached to PRGs and a brief description of the main classes. As mentioned above, there is a dropdown tab where users can choose between the reference resistance gene, putative resistance gene, Avr gene or all resistance gene pages. Clicking on one of these options displays a list of all those type of genes in the database. Hence, by clicking on one of these genes, users will be able to reach the gene details page. The gene reference detail page and the gene putative detail page are composed of the same five sections: gene description, gene information, gene model, BLAST search results and gene sequence. In the gene information section, there is a table containing the general features of a gene. In the gene model section (Figure 2C), users will be able to explore the resistance domains of a gene in an interactive way. With regard to the Avr gene details page, it must be mentioned that in this case there are only three sections: gene description, gene information and gene sequence.
The ‘Pathogens’ page shows a list of all the pathogens stored in this database. In the pathogen description section (Figure 2D), information about that specific pathogen is displayed such as other names, its NCBI accession number or a photo. In the Avr section, a table which contains a list of all known Avr genes belonging to this pathogen is shown. Finally, in the list of diseases section, a table which contains a list of all known plant diseases caused by this pathogen is displayed.
Finally, the ‘Diseases’ page shows a list of all diseases stored in this database and their description, and the disease detail page is only composed of two sections: disease description and presence among species.
DRAGO tool implementation and InterProScan prediction comparison
Version 2 of the DRAGO pipeline was created and tested in the A. thaliana proteome. Our tool was able to identify >1700 putative PRGs in said dataset. The protein sequences of these genes were also analyzed by InterProScan, thus allowing a comparison between both tools. The results showed that 93.98% of the putative pathogen recognition protein sequences had a perfect match between the putative PR-domains predicted by both tools whereas 97.93% of these sequences had a perfect match or had more predicted PR-domains by DRAGO 2, suggesting that DRAGO 2 might be more sensitive. On the other hand, if the detection of the CC domains is not included in the analysis, the number of putative pathogen recognition protein sequences with a perfect match or more putative PR-domains predicted by DRAGO 2 increases up to 99.21%.
Tool updates: DRAGO 2 tool and BLAST search
DRAGO 2 was integrated in the PRGdb web portal. Users can make use of it to predict putative PR-domains in their own sequences, whether DNA or protein. DRAGO 2 screening is performed through an intuitive web-based interface analysis suitable for non-experienced users.
By clicking on the DRAGO 2 option, users will be able to reach the DRAGO 2 form page. This form contains a unique box to paste the input sequences in FASTA format.
By clicking on the BLAST option, users will be able to reach the BLAST form page. This form contains a selection box to choose the PRGs target database (putative, reference or both), a second selection box to choose the analysis required (either blastp or blastx), a box to set the E-value filter and finally another box to paste the input sequences in FASTA format.
New annotations and data
On the one hand, the extensive bibliographic search yielded 41 new cloned PR genes from different resistance families (see the list of these genes and references in Supplementary Table S2). This means a total number of 153 reference genes were stored in the new database. On the other hand, DRAGO 2 was executed to predict putative pathogen recognition genes and to arrange them into PRG classes. More than 177 000 putative PRGs have been classified and stored in PRGdb 3.0. CTN, CTNK, CNLK, CTLK, TNLK, CTNLK, CC-only and transmembrane-only classes have been excluded from the results since none of the species showed these domain combinations. The highest number of predicted PRG proteins in all analysed proteomes belong to RLK, RLP and N with 23 211, 17 143 and 12 495 predicted PRGs respectively. Very rare combinations at low frequencies were also found, such as CTK, TLK and TNK, with a count of 1, 1 and 4, respectively. In contrast, other unknown domain combinations reached an unexpected frequency, with CK and CLK reaching a count of 9099 and 529 respectively.
Additionally, all putative and reference pathogen recognition genes were pooled together and blasted (blasp 2.2.30+) against themselves with parameters -max_hsps_per_subject 1 and -E-value 1e-06. The output results are displayed on the website in the gene details section.
CONCLUSION/FUTURE DIRECTIONS
In recent years, the availability of large scale data is rapidly increasing. The ability of investigators to use this data meaningfully is highly dependent on efficient technology search and data management that is best housed in a community resource such as PRGdb. We will continue to incorporate new data as they become available, including sequence and gene expression data from large-scale genomics projects. Several groups are planning to share manually curated PRG annotations in different species and transcriptomic data related to pathogen plant response. Finally, we have established a collaboration to capture more pathogen related data as well. For all these reason we think that PRGdb v3 could be a reference for PRGs studies.
AVAILABILITY
PRGdb v3 is available at http://prgdb.org.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge Lugi Faino for his help on Avr genes, Toni Hermoso Pulido for the database migration from PRGdb v2 and Freddy Monteiro for his feedback on the web interface.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Università degli Studi di Napoli Federico II; Sequentia Biotech SL. Funding for open access charge: Department of Soil, Plant, Environment and Animal Production Sciences, University of Naples ‘Federico II’; Sequentia Biotech SL.
Conflict of interest statement. None declared.
REFERENCES
- 1. Prohens J. Plant breeding: a success story to be continued thanks to the advances in genomics. Front. Plant Sci. 2011; 2:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. FAO Plants vital to human diets but face growing risks from pests and diseases. 2016; http://www.fao.org/news/story/en/item/409158/icode/.
- 3. Piquerez S.J.M., Harvey S.E., Beynon J.L., Ntoukakis V.. Improving crop disease resistance: lessons from research on arabidopsis and tomato. Front. Plant Sci. 2014; 5:671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andolfo G., Ercolano M.R.. Plant innate immunity multicomponent model. Front. Plant Sci. 2015; 6:987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ercolano M.R., Sanseverino W., Carli P., Ferriello F., Frusciante L.. Genetic and genomic approaches for R-gene mediated disease resistance in tomato: retrospects and prospects. Plant Cell Rep. 2012; 31:973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pérez-de-Castro A.M., Vilanova S., Cañizares J., Pascual L., Blanca J.M., Díez M.J., Prohens J., Picó B.. Application of genomic tools in plant breeding. Curr. Genomics. 2012; 13:179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M. et al. The arabidopsis information resource (TAIR): a model organism database providing a centralized, curated gateway to arabidopsis biology, research materials and community. Nucleic Acids Res. 2003; 31:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fernandez-Pozo N., Menda N., Edwards J.D., Saha S., Tecle I.Y., Strickler S.R., Bombarely A., Fisher-York T., Pujar A., Foerster H. et al. The Sol Genomics Network (SGN)-from genotype to phenotype to breeding. Nucleic Acids Res. 2015; 43:D1036–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fei Z., Tang X., Alba R., Giovannoni J.. Tomato Expression Database (TED): a suite of data presentation and analysis tools. Nucleic Acids Res. 2006; 34:D766–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Proost S., Van Bel M., Vaneechoutte D., Van De Peer Y., Inzé D., Mueller-Roeber B., Vandepoele K.. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 2015; 43:D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanseverino W., Hermoso A., D’Alessandro R., Vlasova A., Andolfo G., Frusciante L., Lowy E., Roma G., Ercolano M.R.. PRGdb 2.0: towards a community-based database model for the analysis of R-genes in plants. Nucleic Acids Res. 2013; 41:D1167–D1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40:1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L. et al. Ensembl 2016. Nucleic Acids Res. 2016; 44:D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2012; 41:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004; 5:113–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson L.S., Eddy S.R., Portugaly E.. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010; 11:431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bateman A., Martin M.J., O’Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R. et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lupas A., Van Dyke M., Stock J.. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996; 266:513–525. [DOI] [PubMed] [Google Scholar]
- 20. Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 2001; 305:567–580. [DOI] [PubMed] [Google Scholar]
- 21. Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.