Abstract
The National Center for Biotechnology Information (NCBI) provides a large suite of online resources for biological information and data, including the GenBank® nucleic acid sequence database and the PubMed database of citations and abstracts for published life science journals. The Entrez system provides search and retrieval operations for most of these data from 39 distinct databases. The E-utilities serve as the programming interface for the Entrez system. Augmenting many of the Web applications are custom implementations of the BLAST program optimized to search specialized data sets. New resources released in the past year include PubMed Data Management, RefSeq Functional Elements, genome data download, variation services API, Magic-BLAST, QuickBLASTp, and Identical Protein Groups. Resources that were updated in the past year include the genome data viewer, a human genome resources page, Gene, virus variation, OSIRIS, and PubChem. All of these resources can be accessed through the NCBI home page at www.ncbi.nlm.nih.gov.
INTRODUCTION
NCBI overview
The National Center for Biotechnology Information (NCBI), a center within the National Library of Medicine at the National Institutes of Health, was created in 1988 to develop information systems for molecular biology. Since the beginning the foundation of these systems has been molecular sequence data, such as the nucleic acid sequence data in GenBank® (1), which NCBI continues to maintain and which continues to receive data through the international collaboration with the DNA Data Bank of Japan (DDBJ) and the European Nucleotide Archive (ENA) as well as from the scientific community. Over the years the amount and variety of data that NCBI maintains has expanded enormously, and can be generally divided into six categories: Literature, Health, Genomes, Genes, Proteins, and Chemicals (Table 1). Each of these six categories has a corresponding web page that lists the relevant databases and tools, along with links to tutorials and other information. Links to these pages are also provided in Table 1. NCBI also provides a variety of services to support the research enterprise: (i) facilities that allow submission of scientific data and open-access publications, (ii) facilities for downloading large and/or customized datasets, (iii) educational events and materials about NCBI products, (iv) software and services to support an expanding developer community, (v) software tools to analyze and/or display NCBI data, and 6) direct involvement in research in computational biology. These services, along with all other data resources, are available through the NCBI home page at www.ncbi.nlm.nih.gov. In most cases, the data underlying these resources and executables for the software described are available for download at ftp.ncbi.nlm.nih.gov.
Table 1. The Entrez Databases (as of 11 September 2017).
| Database | Records | Annual Growth | Description |
|---|---|---|---|
| Literature | www.ncbi.nlm.nih.gov/home/literature.shtml | ||
| PubMed Central | 4 527 796 | 11.35% | full-text journal articles |
| Books | 584 666 | 10.70% | books and reports |
| PubMed | 27 575 666 | 4.40% | scientific and medical abstracts/citations |
| MeSH | 272 224 | 2.58% | ontology used for PubMed indexing |
| NLM Catalog | 1 568 517 | 1.08% | index of NLM collections |
| Health | www.ncbi.nlm.nih.gov/home/health.shtml | ||
| ClinVar | 329 260 | 106.84% | human variations of clinical significance |
| dbGaP | 260 869 | 16.64% | genotype/phenotype interaction studies |
| PubMed Health | 69 322 | 10.05% | clinical effectiveness, disease and drug reports |
| GTR | 52 685 | 8.38% | genetic testing registry |
| MedGen | 296 696 | 1.49% | medical genetics literature and links |
| Genomes | www.ncbi.nlm.nih.gov/home/literature.shtml | ||
| Genome | 25 433 | 49.94% | genome sequencing projects by organism |
| SRA | 4 464 466 | 44.37% | high-throughput DNA and RNA sequence read archive |
| Assembly | 128 427 | 41.55% | genome assembly information |
| BioSample | 6 921 928 | 32.50% | descriptions of biological source materials |
| SNP | 1 070 203 043 | 30.62% | short genetic variations |
| BioProject | 246 934 | 27.30% | biological projects providing data to NCBI |
| Nucleotide | 244 630 453 | 16.41% | DNA and RNA sequences |
| Taxonomy | 1 752 913 | 8.38% | taxonomic classification and nomenclature catalog |
| dbVar | 6 571 714 | 6.89% | genome structural variation studies |
| GSS | 39 972 895 | 0.90% | genome survey sequences |
| Clone | 38 325 051 | 0.63% | genomic and cDNA clones |
| Probe | 32 406 650 | 0.01% | sequence-based probes and primers |
| BioCollections | 7 246 | N/A* | culture collections, museums, and herbaria |
| Genes | www.ncbi.nlm.nih.gov/home/genes.shtml | ||
| Gene | 29 441 891 | 20.90% | collected information about gene loci |
| GEO DataSets | 2 299 715 | 14.51% | functional genomics studies |
| PopSet | 281 116 | 9.25% | sequence sets from phylogenetic and population studies |
| EST | 76 444 005 | 0.25% | expressed sequence tag sequences |
| GEO Profiles | 128 414 055 | -.- | gene expression and molecular abundance profiles |
| UniGene | 6 473 284 | -.- | clusters of expressed transcripts |
| HomoloGene | 141 268 | -.- | homologous gene sets for selected organisms |
| Proteins | www.ncbi.nlm.nih.gov/home/proteins.shtml | ||
| Protein | 430 457 706 | 39.85% | protein sequences |
| Structure | 132 219 | 8.86% | experimentally-determined biomolecular structures |
| Conserved Domains | 56 066 | 6.97% | conserved protein domains |
| Protein Clusters | 820 546 | 0.00% | sequence similarity-based protein clusters |
| Identical Protein Groups | 141 763 601 | N/A | groups of identical protein sequences |
| Chemicals | www.ncbi.nlm.nih.gov/home/chemicals.shtml | ||
| BioSystems | 983 968 | 11.82% | molecular pathways with links to genes, proteins, and chemicals |
| PubChem Substance | 229 830 586 | 2.99% | deposited substance and chemical information |
| PubChem BioAssay | 1 252 796 | 2.80% | bioactivity screening studies |
| PubChem Compound | 91 752 585 | 0.08% | chemical information with structures, information, and links |
*Database was first released in 2017
This article provides a brief overview of the NCBI Entrez system of databases, followed by a summary of resources that were either introduced or significantly updated in the past year. More complete discussions of NCBI resources can be found on the home pages of individual databases, on the NCBI Learn page (www.ncbi.nlm.nih.gov/learn/), or in the NCBI Handbook (www.ncbi.nlm.nih.gov/books/NBK143764/).
The Entrez system
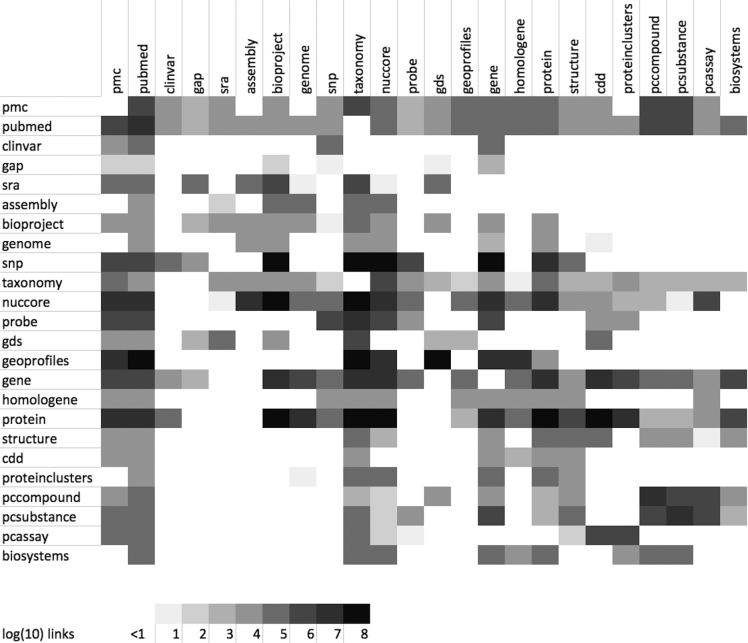
Entrez (2) is an integrated database retrieval system that provides access to a diverse set of 39 databases that together contain 2.5 billion records (Table 1). Links to the web portal for each of these databases are provided on the Entrez GQuery page (www.ncbi.nlm.nih.gov/gquery/). Entrez supports text searching using simple Boolean queries, downloading of data in various formats, and linking records between databases based on asserted relationships. In their simplest form, these links may be cross-references between a sequence and the abstract of the paper in which it is reported, or between a protein sequence and either its coding DNA sequence or its 3D-structure. Computationally derived links between neighboring records, such as those based on computed similarities among PubMed abstracts, allow rapid access to groups of related records. A summary of available links for selected databases is shown in Figure 1. The LinkOut service expands the range of links to include external resources, such as organism-specific genome databases. The records retrieved in Entrez can be displayed in many formats and downloaded singly or in batches. An Application Programming Interface for Entrez functions (the E-utilities) is available, and detailed documentation is provided at eutils.ncbi.nlm.nih.gov.
Figure 1.
Graphical Depiction of Selected Entrez Links. Each cell in the matrix is shaded according to the log (base 10) of the number of records in the source database (rows) that have an Entrez link to the destination database (columns). Diagonal cells represent computational links (e.g. pubmed related articles) and off-diagonal cells assert biological relationships (e.g. nuccore to taxonomy). The matrix is not diagonal because an individual record in a source database may have many links to a destination database (e.g. genome to protein).
Data sources and collaborations
NCBI receives data from three sources: direct submissions from researchers, national and international collaborations or agreements with data providers and research consortia, and internal curation efforts. One notable effort is the Genome Reference Consortium (GRC) that provides the reference genome assemblies for human, mouse, zebrafish, and chicken (www.ncbi.nlm.nih.gov/grc/). Details about direct submission processes are available from the NCBI Submit page (www.ncbi.nlm.nih.gov/home/submit.shtml) and from the resource home pages (e.g. the GenBank page, www.ncbi.nlm.nih.gov/genbank/). NCBI staff provide identifiers to submitters for their data generally within 2–5 business days, depending on the destination database and the complexity of the submission. More information about the various collaborations, agreements, and curation efforts are also available through the home pages of the individual resources.
RECENT DEVELOPMENTS
Literature updates
PubMed licensing
In the past, NLM offered downloads of the PubMed dataset after signing a free license agreement. This policy has now changed, and the entire PubMed dataset is now available under certain terms and conditions but without a signed license. Each December, NLM releases a complete baseline dataset in XML (ftp.ncbi.nlm.nih.gov/pubmed/baseline/). Thereafter, an update file is released each day that contains new, revised, and deleted records (ftp.ncbi.nlm.nih.gov/pubmed/updatefiles/). More details are available in this README file: ftp://ftp.ncbi.nlm.nih.gov/pubmed/updatefiles/README.txt.
PubMed data management (PMDM)
One of the most common maintenance tasks in PubMed is correcting citation errors, such as author names, affiliations, or article titles. In the past, users reported such errors to NLM who then worked with publishers to correct these problems. To improve this, in late 2016 NLM released the PubMed Data Management system, an online service that allows publishers to correct their PubMed data directly. Once entered, corrections appear on PubMed in 24–48 h. As a result, users can now report PubMed citation errors directly to the relevant publisher, who can then make the corrections. Generally, these improvements have significantly simplified and accelerated the correction process.
Genome updates
Sequence identifiers
As described elsewhere (1), NCBI is phasing out the practice of assigning GI numbers as identifiers for records in the sequence databases (Nucleotide, EST, GSS, Popset and Protein). Instead, the accession.version is now the primary and consistent identifier for all sequence records at NCBI. As time progresses, more new sequence records will be assigned only accession.version identifiers (current examples of such sequences are unannotated WGS and TSA contigs). In November 2016, NCBI removed GI numbers from GenBank and FASTA formats obtained from the web, APIs, or FTP (they will remain in XML and ASN.1 formats). In addition, the E-utilities now accept accession.version identifiers as input and can provide them as output when the parameter idtype is set to ‘acc’.
RefSeq functional elements
In May 2017 NCBI released a new dataset of human and mouse RefSeq Functional Elements (https://www.ncbi.nlm.nih.gov/refseq/functionalelements/). This new dataset, which supplements NCBI’s conventional gene representations, is a curated set of non-genic functional elements from human and mouse that have been experimentally validated in the literature. It includes genomic elements that function in gene regulation, chromosome organization, recombination, and DNA replication. Each functional element is represented both as a genomic RefSeq record (in GenBank flat file and ASN.1 formats) and also as a new ‘biological region’ type of Gene record. These Gene records contain metadata about the element, details about its genome annotation, associated literature, and more. An example functional element is the locus control region for human beta-globin, which has Gene ID 109580095, gene symbol HBB-LCR, and sequence NG_052895. These records are all linked to BioProject PRJNA343958 and can be downloaded from that resource as well as from the RefSeq, Genomes, and Gene FTP sites. In addition to its general-purpose use in basic biological research, this dataset is expected to be highly useful for interpreting disease-associated variation in non-genic regions.
Genome data viewer
The NCBI Genome Data Viewer (GDV) has a new home page that allows users to explore the more than 100 eukaryotic genomes that GDV supports (www.ncbi.nlm.nih.gov/genome/gdv/). The page displays an interactive taxonomic tree that organizes these genomes, and clicking a leaf of the tree updates a panel with statistics and links for the represented genome. These panels provide easy access to displays in GDV along with the genome's BLAST interface, and also show graphical views of the individual chromosomes. The panels also support searches in the genome by gene symbol, location, and phenotype, and these searches lead to views in GDV.
Human genome resources
To centralize various resources that support the human genome, NCBI has released a new Human Genome Resources page (www.ncbi.nlm.nih.gov/genome/guide/human). In addition to providing a search interface for the human genome along with a graphical depiction of the chromosomes that lead to views in GDV, the page organizes content in several sections: Download, Browse, View, and Learn. The page provides several downloads for both the current (GRCh38) and previous (GRCh37) human genome builds, links to over 20 related tools and resources, several webinars and video tutorials, and over ten fact sheets.
Genome data download
The Assembly database, which catalogs genomic datasets from both GenBank and RefSeq, now includes a control that provides easy downloads of these entire datasets from a web browser. After conducting a search in Assembly, a Download Assemblies button will appear at the top of the results that opens a dialog allowing the selected assemblies (or all by default) to be downloaded in many different formats. Example formats include FASTA, GFF3, and GenBank flat files as well as statistics and feature tables.
Variation services APIs
NCBI now offers a new set of APIs for comparing and grouping genomic variants (api.ncbi.nlm.nih.gov/variation/v0/). These services use a novel data model named SPDI (Sequence-ID, Position, Deleted sequence, Inserted sequence), and can convert both HGVS and VCF data formats into SPDI (www.ncbi.nlm.nih.gov/variation/notation/). One major advantage of this new model is that it allows users to input several HGVS and/or VCF variants and determine if these refer to the same variant. They also can return any RefSNP IDs that correspond to the input variant. More details are available in the documentation (www.ncbi.nlm.nih.gov/variation/services/).
Virus variation
NCBI’s resources devoted to variations in viral genomes have a new interface that improves search and retrieval for seven viral subgroups: Influenza, Dengue, Zika, Rotavirus, West Nile, MERS coronavirus, and Ebolavirus (3). Each subgroup has a dedicated module that supports searches using standardized gene and protein names as well as sample information. Searches can be limited to full length sequences, and identical sequences can be collapsed to clarify the results. Once a set of sequences is retrieved, the tool can use them to create a multiple sequence alignment, build a tree, or download the data.
OSIRIS
The Open Source Independent Review and Interpretation Software is a powerful standalone quality assurance tool for the assessment of multiplex short tandem repeat profile (STR) data (https://www.ncbi.nlm.nih.gov/projects/SNP/osiris/). This application can be installed locally for rapid analysis of STR profiles used in clinical monitoring of stem cell transplants, identifying tissue samples, and verifying cell lines. OSIRIS was updated in 2017 to increase the discrimination of artifacts, the accuracy of analysis, and the overall usability. Users can download OSIRIS source code at github.com/ncbi/osiris.
Gene updates
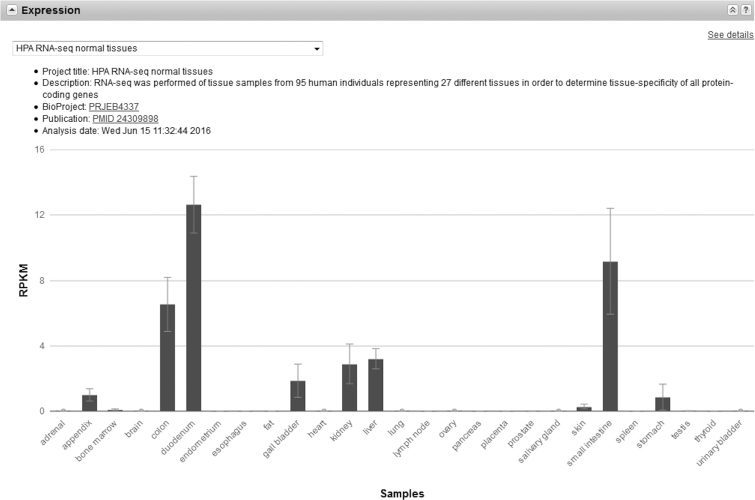
The Gene resource now includes representative expression profiles, both as a graphical representation of each gene's expression integrated into its full report page (see Figure 2), and as datasets available for download. Expression profiles are both useful complements to already characterized gene functions and also potential means of initially characterizing the function of novel genes (4,5). Initial datasets are currently available for human, mouse, and rat. In the future a text summary will accompany each gene's expression profile, and these data will be indexed within the Entrez query system. These expression profiles are computed from RNA-seq alignments generated by NCBI’s eukaryotic genome annotation pipeline. This process selects representative datasets publicly available in SRA based on their breadth of tissue and developmental samples, their read characteristics, and other considerations. After aligning reads from a sample to the genomic sequence, for each gene the read coverage is computed (compared to all annotated exons for that gene), normalized to all reads aligned to the genome, and used to derive reads per kilobase per million reads placed (RPKM) across the gene. Data from biological replicates within the same SRA project are averaged and reported with the standard deviation. Expression levels from different SRA projects are reported independently, given the lack of clear standards for coping with batch effects from heterogeneous sample preparations (6,7).
Figure 2.
Expression profile data for human TRIM15 (gene ID 89870) as displayed on the Gene full report (www.ncbi.nlm.nih.gov/gene/89870).
BLAST updates
Magic-BLAST
Magic-BLAST is a command-line tool that maps large sets of next-generation RNA or DNA sequencing data against a whole genome or transcriptome. Unlike typical BLAST, Magic-BLAST optimizes alignments based on a composite score for a read pair, summing the score of all exons in the case of RNA-seq data. An entire next generation run serves as the query, and can be provided as an SRA accession or as data in SRA, FASTA, FASTQ, or FASTC formats. It is preferable that the reference genome or transcriptome be provided as a BLAST database, and procedures for constructing these, along with other details, are provided on the Magic-BLAST FTP site (ftp.ncbi.nlm.nih.gov/blast/executables/magicblast/).
QuickBLASTp
QuickBLASTp is a new, accelerated protein BLAST algorithm that performs a rapid k-mer search against the nr database. This k-mer search uses a word size of 5 and is tuned so that it returns ∼97% of sequences with >70% sequence identity to the query, and ∼98% of sequences with >80% identity. This algorithm works best for queries longer than 50 residues, and is limited to queries of <10 000 residues. Access to QuickBLASTp is provided as an algorithm option on the main BLASTp web page.
SmartBLAST
The SmartBLAST service quickly returns the most similar proteins to a query, and was updated in 2017 to prioritize matching proteins to the ‘landmark’ database that contains proteomes from 26 well-annotated genomes. The upper panel of the display now shows the top five matches from the landmark database, while additional matches from nr are listed in a separate panel. In addition, the results page shows more information about Conserved Domain records in the query, thereby assisting in identifying the functional elements conserved between the query and matching sequences.
Protein updates
In 2014 NCBI introduced the ‘Identical Protein Report’ to the Protein database to clarify the relationships between WP sequences and the set of individual Nucleotide CDS sequences they represent (8). Now these reports have been improved and collected in a new resource called Identical Protein Groups (www.ncbi.nlm.nih.gov/ipg/). This IPG resource includes all NCBI protein sequences, including records from INSDC, RefSeq, Swiss-Prot, and PDB, with links to nucleotide coding sequences from GenBank and RefSeq. The title of each record is derived from the ‘best’ sequence in each group, where the hierarchy for determining the best sequence is RefSeq > Swiss-Prot > PIR, PDB > GenBank > patent (www.ncbi.nlm.nih.gov/ipg/docs/faq/). Searches in this database can be filtered by database source, taxonomy, and the number of sequences in the group. These reports continue to be available through the E-utility EFetch with &db = protein&rettype = ipg (eutils.ncbi.nlm.nih.gov).
Chemical updates
PubChem (9,10) is a resource that provides information on various chemical entities, including small molecules, siRNAs, miRNAs, carbohydrates, lipids, peptides, chemically modified macromolecules, and many others. In the past year, PubChem introduced several major improvements. One was a new version of PubChem Widgets (Widgets 2.0f) that enables web developers to display PubChem content on their own webpages. Widgets 2.0f provides many additional data views, simplifies the process of embedding widgets, and makes it easier for a developer to resize widgets, allowing more adaptability to different screen sizes.
The PubChem Data Sources page summarizes data contributors to PubChem, and this page was updated to provide new and improved capabilities to navigate as a function of data type, category, and country, while also including keyword searching, counts, and geographic visualization. In addition, the Data Sources page makes it easier to separate active data contributors from non-active (‘legacy’) data contributors.
Molecular weights in PubChem were updated using the latest International Union of Pure and Applied Chemistry (IUPAC) recommendations for atomic mass and isotopic composition information (11,12). Increasingly the scientific community is recognizing complex issues with average atomic weight and isotopic data, as greater degrees of precision in atomic masses and variations in isotopic abundance are known. PubChem now uses the ‘conventional atomic weights’ described by IUPAC when available. In addition, PubChem is now restricting the allowed isotopes for a given element to those with a half-life of one millisecond or greater.
The PubChem resource introduced Target Summary pages in 2017 that collect bioactivity data about particular genes. These pages are available for any BioAssay record that has a protein or gene target, and are linked from the record's ‘BioAssay Target’ section. Target Summary pages contain information about the protein targets encoded by the gene, known drugs and other compounds tested against these targets, other BioAssays that involve these targets, and a variety of other biological information about the gene. Additional details about these and other PubChem developments are available on the PubChem blog (pubchemblog.ncbi.nlm.nih.gov).
FOR FURTHER INFORMATION
The resources described here include documentation, other explanatory material, and references to collaborators and data sources on their respective web sites. An alphabetical list of NCBI resources is available from a link above the category list on the left side of the NCBI home page. The NCBI Help Manual and the NCBI Handbook (www.ncbi.nlm.nih.gov/books/NBK143764/), both available as links in the common page footer, describe the principal NCBI resources in detail. The NCBI Learn page (www.ncbi.nlm.nih.gov/learn/) provides links to documentation, tutorials, webinars, courses, and upcoming conference exhibits. A variety of video tutorials are available on the NCBI YouTube channel that can be accessed through links in the standard NCBI page footer. A user-support staff is available to answer questions at info@ncbi.nlm.nih.gov, and users can view support articles at support.ncbi.nlm.nih.gov. Updates on NCBI resources and database enhancements are described on the NCBI Insights blog (ncbiinsights.ncbi.nlm.nih.gov), NCBI social media sites (FaceBook, Twitter, and LinkedIn), and the several mailing lists and RSS feeds that provide updates on services and databases. Links to these resources are in the NCBI page footer and on NCBI Insights.
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
APPENDIX
NCBI Resource Coordinators: Richa Agarwala, Tanya Barrett, Jeff Beck, Dennis A Benson, Colleen Bollin, Evan Bolton, Devon Bourexis, J Rodney Brister, Stephen H Bryant, Kathi Canese, Mark Cavanaugh, Chad Charowhas, Karen Clark, Ilya Dondoshansky, Michael Feolo, Lawrence Fitzpatrick, Kathryn Funk, Lewis Y Geer, Viatcheslav Gorelenkov, Alan Graeff, Wratko Hlavina, Brad Holmes, Mark Johnson, Brandi Kattman, Viatcheslav Khotomlianski, Avi Kimchi, Michael Kimelman, Masato Kimura, Paul Kitts, William Klimke, Alex Kotliarov, Sergey Krasnov, Anatoliy Kuznetsov, Melissa J Landrum, David Landsman, Stacy Lathrop, Jennifer M Lee, Carl Leubsdorf, Zhiyong Lu, Thomas L Madden, Aron Marchler-Bauer, Adriana Malheiro, Peter Meric, Ilene Karsch-Mizrachi, Anatoly Mnev, Terence Murphy, Rebecca Orris, James Ostell, Christopher O'Sullivan, Vasuki Palanigobu, Anna R Panchenko, Lon Phan, Borys Pierov, Kim D Pruitt, Kurt Rodarmer, Eric W Sayers, Valerie Schneider, Conrad L Schoch, Gregory D Schuler, Stephen T Sherry, Karanjit Siyan, Alexandra Soboleva, Vladimir Soussov, Grigory Starchenko, Tatiana A Tatusova, Francoise Thibaud-Nissen, Kamen Todorov, Bart W Trawick, Denis Vakatov, Minghong Ward, Eugene Yaschenko, Aleksandr Zasypkin, Kerry Zbicz.
Contributor Information
NCBI Resource Coordinators:
Richa Agarwala, Tanya Barrett, Jeff Beck, Dennis A Benson, Colleen Bollin, Evan Bolton, Devon Bourexis, J Rodney Brister, Stephen H Bryant, Kathi Canese, Mark Cavanaugh, Chad Charowhas, Karen Clark, Ilya Dondoshansky, Michael Feolo, Lawrence Fitzpatrick, Kathryn Funk, Lewis Y Geer, Viatcheslav Gorelenkov, Alan Graeff, Wratko Hlavina, Brad Holmes, Mark Johnson, Brandi Kattman, Viatcheslav Khotomlianski, Avi Kimchi, Michael Kimelman, Masato Kimura, Paul Kitts, William Klimke, Alex Kotliarov, Sergey Krasnov, Anatoliy Kuznetsov, Melissa J Landrum, David Landsman, Stacy Lathrop, Jennifer M Lee, Carl Leubsdorf, Zhiyong Lu, Thomas L Madden, Aron Marchler-Bauer, Adriana Malheiro, Peter Meric, Ilene Karsch-Mizrachi, Anatoly Mnev, Terence Murphy, Rebecca Orris, James Ostell, Christopher O'Sullivan, Vasuki Palanigobu, Anna R Panchenko, Lon Phan, Borys Pierov, Kim D Pruitt, Kurt Rodarmer, Eric W Sayers, Valerie Schneider, Conrad L Schoch, Gregory D Schuler, Stephen T Sherry, Karanjit Siyan, Alexandra Soboleva, Vladimir Soussov, Grigory Starchenko, Tatiana A Tatusova, Francoise Thibaud-Nissen, Kamen Todorov, Bart W Trawick, Denis Vakatov, Minghong Ward, Eugene Yaschenko, Aleksandr Zasypkin, and Kerry Zbicz
REFERENCES
- 1. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2017; 45:D37–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schuler G.D., Epstein J.A., Ohkawa H., Kans J.A.. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996; 266:141–162. [DOI] [PubMed] [Google Scholar]
- 3. Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., Schaffer A.A., Brister J.R.. Virus Variation Resource - improved response to emergent viral outbreaks. Nucleic Acids Res. 2017; 45:D482–D490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ozsolak F., Milos P.M.. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011; 12:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fagerberg L., Hallstrom B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteomics. 2014; 13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu J., Gong B., Wu L., Thakkar S., Hong H., Tong W.. Comprehensive assessments of RNA-seq by the SEQC Consortium: FDA-led efforts advance precision medicine. Pharmaceutics. 2016; 8, doi:10.3390/pharmaceutics8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ma S., Sung J., Magis A.T., Wang Y., Geman D., Price N.D.. Measuring the effect of inter-study variability on estimating prediction error. PLoS One. 2014; 9:e110840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014; 42:D7–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y., Bryant S.H., Cheng T., Wang J., Gindulyte A., Shoemaker B.A., Thiessen P.A., He S., Zhang J.. PubChem BioAssay: 2017 update. Nucleic Acids Res. 2017; 45:D955–D963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meija J., Coplen T.B., Berglund M., Brand W.A., De Bievre P., Groning M., Holden N.E., Irrgeher J., Loss R.D., Walczyk T. et al. Atomic weights of the elements 2013 (IUPAC Technical Report). Pure Appl. Chem. 2016; 88:265–291. [Google Scholar]
- 12. Meija J., Coplen T.B., Berglund M., Brand W.A., De Bievre P., Groning M., Holden N.E., Irrgeher J., Loss R.D., Walczyk T. et al. Isotopic compositions of the elements 2013 (IUPAC Technical Report). Pure Appl. Chem. 2016; 88:293–306. [Google Scholar]