Abstract
Recent cancer pharmacogenomic studies profiled large panels of cell lines against hundreds of approved drugs and experimental chemical compounds. The overarching goal of these screens is to measure sensitivity of cell lines to chemical perturbations, correlate these measures to genomic features, and thereby develop novel predictors of drug response. However, leveraging these valuable data is challenging due to the lack of standards for annotating cell lines and chemical compounds, and quantifying drug response. Moreover, it has been recently shown that the complexity and complementarity of the experimental protocols used in the field result in high levels of technical and biological variation in the in vitro pharmacological profiles. There is therefore a need for new tools to facilitate rigorous comparison and integrative analysis of large-scale drug screening datasets. To address this issue, we have developed PharmacoDB (pharmacodb.pmgenomics.ca), a database integrating the largest cancer pharmacogenomic studies published to date. Here, we describe how the curation of cell line and chemical compound identifiers maximizes the overlap between datasets and how users can leverage such data to compare and extract robust drug phenotypes. PharmacoDB provides a unique resource to mine a compendium of curated cancer pharmacogenomic datasets that are otherwise disparate and difficult to integrate.
INTRODUCTION
Cancer has emerged as one of the principal causes of mortality in the 21st century (1). It is a collection of related diseases with widely different prognosis and response to therapy (2). This heterogeneity poses challenges for treatment, as patients with the same diagnosis often have different responses to treatment and may develop resistances at different rates (3). The genesis, progression, and response to pharmacotherapy of cancer is largely determined by the molecular state and features of the tumor cells (4). This observation spurred the development of high-throughput pharmacogenomics studies to investigate the relationships between genomic, transcriptomic and proteomic features of cancer cells and their response to treatment with small molecule compounds.
Immortalized cancer cell lines are the most widely-used models to study response of tumors to anticancer compounds (5). In addition to being comprehensively profiled at the molecular level, cancer cell lines can be cultured to conduct high-throughput drug screening studies, where large panels of compounds are screened for their efficacy of halting the growth or killing molecularly distinct cancer tumor models (6). Over the past decade, several large studies combining high-throughput in vitro drug screening with molecular profiling of cancer cell lines have been published (7–13). Recognizing that the molecular diversity of cancer cannot be faithfully represented by small panels of cell lines, these studies have assembled large panels of hundreds to over a thousand cell lines, and profiled them at the molecular and pharmacological levels. These valuable data have been publicly released via well-established data repositories and institutional websites.
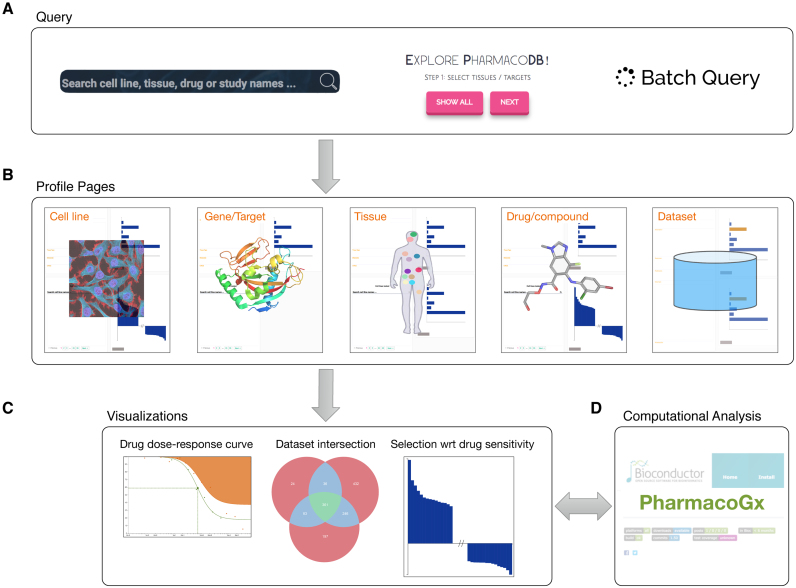
The main limitation of the majority of published cancer pharmacogenomic studies is that they are restricted to the analysis of single datasets. This is primarily due to inconsistent annotations of cell lines and compounds, which prevents direct comparison between datasets (14). Meta-analysis of pharmacogenomic data is further hindered by the lack of standards for statistical modeling of drug dose-response curves and subsequent summarization into drug sensitivity measures (14–17). However, joint analysis of independent datasets holds the potential to improve robustness of research outputs against variations in the complex experimental protocols used in high-throughput drug screening (18). To address these issues we developed PharmacoDB, the first database integrating multiple high-throughput cancer pharmacogenomic datasets (Table 1; Supplementary Figure S1A). PharmacoDB provides an intuitive interface to search and explore these datasets (Figure 1A) based on cell lines and their tissue source, compounds and their targets (Figure 1B), and experiments in which cell viability is measured for cell lines treated with chemical compounds (Figure 1C). Moreover, PharmacoDB provides access to molecular profiles of cell lines and computational analytical tools via linkage to PharmacoGx (Figure 1D; Supplementary Figure S1A), an R/Bioconductor package implementing a suite of statistical modeling functions to jointly analyze molecular features and drug dose-response curves (19). Here, we describe the content of our integrative pharmacogenomic database, the curation process, and its web-interface.
Table 1. Specifications of all the datasets included in PharmacoDB.
| Dataset | # Drugs | # Cells | # Experiments | Viability Assay | Available Molecular Data | Citations |
|---|---|---|---|---|---|---|
| CCLE | 22 | 1061 | 11 670 | CellTiter Glo | mRNA expression, CNV, Mutation | (8) |
| GDSC1000 | 250 | 1109 | 225 480 | Syto60 | mRNA expression, CNV, Mutation, Methylation | (7,13) |
| gCSI | 16 | 754 | 6455 | CellTiter Glo | mRNA expression, CNV, Mutation | (10,11) |
| GRAY | 90 | 84 | 9413 | CellTiter Glo | mRNA expression, CNV, RPPA, Methylation, ExomeSeq | (9,22) |
| FIMM | 52 | 50 | 2561 | CellTiter Glo | None | (12) |
| CTRPv2 | 544 | 888 | 395 263 | CellTiter Glo | None | (20,21) |
| UHNBreast | 4 | 84 | 52 | SRB | RNAseq, RPPA, Mut | (Safikhani et al., accepted, Nat Commun 2017) |
| Total | 759 | 1691 | 650 894 |
Viability assay: Syto60: Proliferation; fluorescent DNA stain (Invitrogen); CellTiter Glo: Viability, membrane integrity, ATP (Promega); SRB: Sulforhodamine B colorimetric. Available molecular data: Mut: targeted mutation data; ExomeSeq: Whole exome-sequencing data; mRNA: gene expression data; methylation: methylation microarray data; CNV: copy number variation data; RPPA: protein expression data using reverse phase protein lysate microarray.
Figure 1.
Main functionalities of PharmacoDB, displaying (A) the interfaces to query the database through searching or exploring available entities, (B) the five primary data types with respective profile pages, (C) the main visualizations of the aggregated data in PharmacoDB and (D) the link to PharmacoGx for extensive computational analysis of pharmacogenomic data.
DATA COLLECTION AND DATABASE CONTENT
Pharmacogenomic studies
PharmacoDB seeks to include the largest published studies investigating the viability response of human cancer cell lines to chemical compound treatment. To date, we have curated seven major cancer studies: The Cancer Cell Line Encyclopedia (CCLE) (8), Genomics of Drug Sensitivity in Cancer (GDSC) (7,13), Genentech Cell Screening Initiative (gCSI) (10,11), the Cancer Therapeutic Response Portal (CTRP) (20,21), the Oregon Health and Science University (OHSU) Breast Cancer Screen by Dr Joe Gray's lab (GRAY) (9,22), the Institute for Molecular Medicine Finland cell viability screen (FIMM) (12), and the University Health Network (Toronto) breast cancer screen (UHNBreast) (Safikhani et al., accepted, Nat Commun 2017). For each study, we downloaded the cell line and compound annotations available with the original publications of the study, either through the journal website or dedicated portals for data sharing made available by the study authors (Table 1; Supplementary Figure S1A; Supplementary Methods).
Annotation of cell lines and chemical compounds
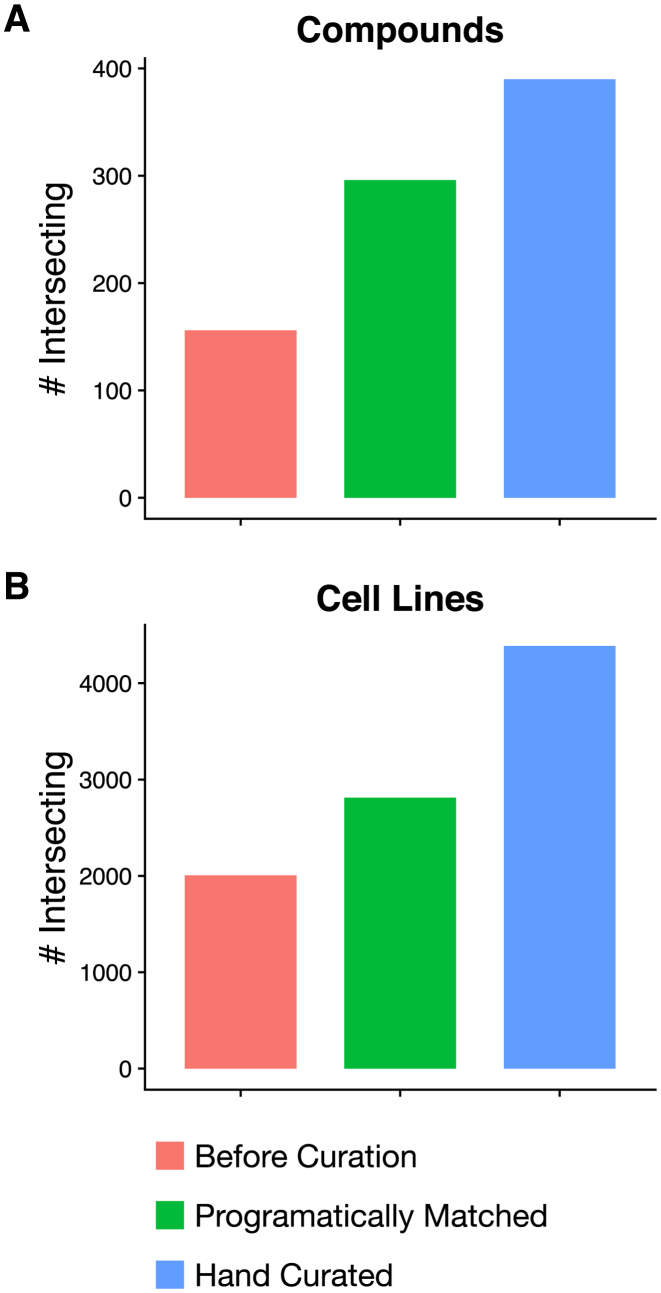
We performed semi-automated curation of all the cell line and compound identifiers with the goal of discovering and maximizing the overlap between the datasets. First, we looked for exact case-insensitive matches of the identifiers used in the dataset undergoing curation to already curated unique identifiers, if applicable. Second, for all remaining compounds and cell lines, a partial, programmatic matching algorithm was used to generate candidate unique identifier matches for each identifier used in the study. These candidate matches were manually reviewed to find the correct match for all compounds and cell lines which had a matching unique identifier. Third, we manually curated the subset of identifiers for which there was no match using the compound and cell line names. For compounds, we used any other provided compound annotations such as the SMILES, InchiKey or PubChem identifier to match them with identifiers available for previously curated compounds. If only the compound name is available, we used the WebChem R package (version 0.2) to query PubChem. If it was possible to retrieve the identifiers of these compounds, then the third step was repeated to find possible matches with previously curated compounds. For cell lines which had no correct matches in the second step, Cellosaurus (23) was queried to generate candidate cell name synonyms, and manual matching was attempted with current unique identifiers. If at the end of these three steps there remained any cell line or compound names that were not matched, a new unique, human interpretable identifier was created based on the name from the dataset currently undergoing curation. This curation process maximized the overlap between datasets (Figure 2; Supplementary Figures S2 and S3). Programmatic matching almost doubled the number of compounds intersecting across datasets while manual curation further increased the intersection by 32% (Figure 2A). Unlike compounds, programmatically matching only moderately increase the intersection across datasets while manual curation was crucial to maximize overlap (Figure 2B). At the dataset level, while some intersections increased only modestly due to similar identifiers being used in the original studies, the benefit was substantial for others. For example, the intersection of compounds tested in GDSC1000 and CTRPv2 more than tripled, from 27 to 90 (Supplementary Figure S2A) and for cell lines, the intersection between CCLE and CTRPv2 quadrupled (Supplementary Figure S2B). While many of the newly matched identifiers differed only in capitalization or hyphenation, computational approaches to mapping identifiers which ignore these differences would be insufficient. These approaches would fail to match certain cases, such as the matching of compound names AZD6244 and Selumetinib, and would also cause mismatches, as for example for the distinct cell lines KMH-2 and KM-H2, which are respectively a Hodgkin's lymphoma cell line and a thyroid gland carcinoma. Differences in naming conventions often create difficulties and confusion for researchers who wish to integrate data from across different studies, and the curation done in PharmacoDB aims to alleviate this barrier to leveraging these valuable pharmacogenomics studies. Overall, we identified 1691 unique cell lines from 41 tissue sources and 759 unique compounds with 673 associated targets.
Figure 2.
The total number of identifier matches between pairs of datasets for (A) cell lines and (B) compounds before curation, after automatically removing differences in capitalization and white space and manually reviewing matches within edit distance of 2, and after undergoing subsequent manual curation.
Annotation of drug targets
To obtain a comprehensive collection of target proteins for the compounds included in PharmacoDB, the union of known drug–target associations from four distinct data sources was integrated into the database. The CTRPv2 study released curated annotations of the protein targets for compounds (20). Additional drug target annotations were retrieved programmatically from the Drug Repurposing Hub (24), DrugBank (25), and ChEMBL (26). For DrugBank, we retrieved the gene symbol for each target using UniProt.ws (version 2.16.0). For ChEMBL, we used the Web API to retrieve gene symbols for the protein targets, subsequently linked to the appropriate GeneCard (27,28).
Comparison to existing databases
Current pharmacogenomic databases focus on cataloguing and curating relationships between genomic variants and the efficacy and/or toxicity of pharmaceutical compounds. PharmGKB currently annotates over 620 drugs with a total of over 18 000 annotations (29). PharmGKB curation includes manual evaluation of the evidence and statistical significance of the gene-compound associations reported in published studies. Those with strong evidence and clinical significance are highlighted for use in clinical practice, while associations with weak evidence or non-significant associations are made available for use in research. PGMD, an effort by Qiagen Bioinformatics, similarly starts with mining the pharmacogenomic literature, but aims to be wider in the scope of genetic variation and additional annotations captured in the database (30). PGMD includes annotations about variants in intergenic regions, and maps each annotation to genomic coordinates, enabling the integration of this data into sequencing studies. This broader scope allowed PGMD to include over 117 000 unique pharmacogenomic annotations encompassing nearly 1400 drugs. The DruGeVar database follows a similar literature mining approach, focusing on FDA and EMA approved drugs which have a pharmacogenomic association on the drug label, leading to a smaller but more clinically actionable database (31).
While PharmGKB, PGMD and DruGeVar rely on literature mining to distill robust, clinically actionable pharmacogenomic associations, PharmacoDB focuses on the massive in vitro pharmacogenomic studies enabled by high-throughput molecular profiling and drug screening assays, which aim to be hypothesis-generating resources for biomarker discovery and drug repurposing. All the phenotypical data included in PharmacoDB are derived from dose–response experiments on cancer immortalized cell lines for approved and experimental compounds, unlike existing databases which mix evidence from preclinical and clinical studies. As such, PharmacoDB is a resource for researchers who have exhausted the existing literature, allowing them to mine existing data to discover evidence related to their compound or gene of interest, and enabling rapid hypothesis generation or in vitro validation.
DATABASE ORGANIZATION AND WEB-INTERFACE
Database implementation
All of the data is stored in a MySQL database running the default MyISAM database engine and with indexing configured on all tables in order to speed up queries (database schema in Supplementary Figure S4). The web interface is implemented using Ruby (version 2.4.1) and Ruby on Rails (version 5). To provide a smooth navigation experience, the front-end is rendered on the server and performance is optimized with use of Turbolinks (version 5.0), which does selective updates and contributes to faster page load times. All charts were produced using d3.js (version 3), a JavaScript library tailored to produce dynamic and interactive data visualizations using SVG, HTML5 and CSS web standards. Every plot generated on PharmacoDB is available for download in the SVG vectorized graphics format and the data used to generate the plot are exportable as spreadsheets.
Search interface
Often, biomedical researchers interested in leveraging pharmacogenomic data are investigating a specific biological question about a given cell line, tissue, drug or target. PharmacoDB is designed to quickly answer the question: ‘What pharmacogenomic data is available for my entity of interest?’. A universal search bar interface allows users to intuitively search across all entities included in the database: datasets, tissues, cell lines, compounds and genes (Figure 1; Supplementary Figure S1). This search bar is enhanced with autocompletion, giving quick feedback to the existence of an entity in our database, and helping with correct spelling and punctuation of entries. The search bar also handles more complex queries, allowing the user to search for pairwise and three way intersections of datasets, and navigate directly to a dose–response curve by querying for a cell line and compound in combination.
Search by synonyms
The same cell line, tissue, or compound entity is often known by several names, which are often used interchangeably in the literature. As described above, semi-manual curation of datasets in PharmacoDB was done to map the synonyms used in each dataset to a unique human interpretable identifier. However, as each user may be more or less familiar with a specific synonym for a given compound or cell line, PharmacoDB was implemented such that it is possible to search for a compound, tissue or cell line by any of the synonyms collected in the curation process. This means that if a researcher is familiar with the name of a compound used in the CTRPv2 dataset, for example, they can use this identifier to find experiments with the same compound across other datasets. The synonyms encountered include different spelling or punctuation as well as completely different names, and enable a more natural interface with the database. Currently, there are 4162 different synonyms used to refer to the 1,691 cell lines, 980 synonyms to refer to the 759 compounds, and 184 synonyms for the 41 tissues in PharmacoDB.
Explore interface
Complementing the Search interface, the explore page serves as a gentle entry point for new users attempting to navigate PharmacoDB. It facilitates discovery of content by presenting to the user all the entities aggregated in the database. User interaction with the explore page occurs in a series of filtering steps to find the entity or experiment of interest. Depending on which selections users make, unrelated annotations are filtered out until they make a selection corresponding to a single query of the database. This will allows the user to quickly navigate through the large collection of entities while having a complete picture of all the targets, tissues, compounds and drugs included in PharmacoDB.
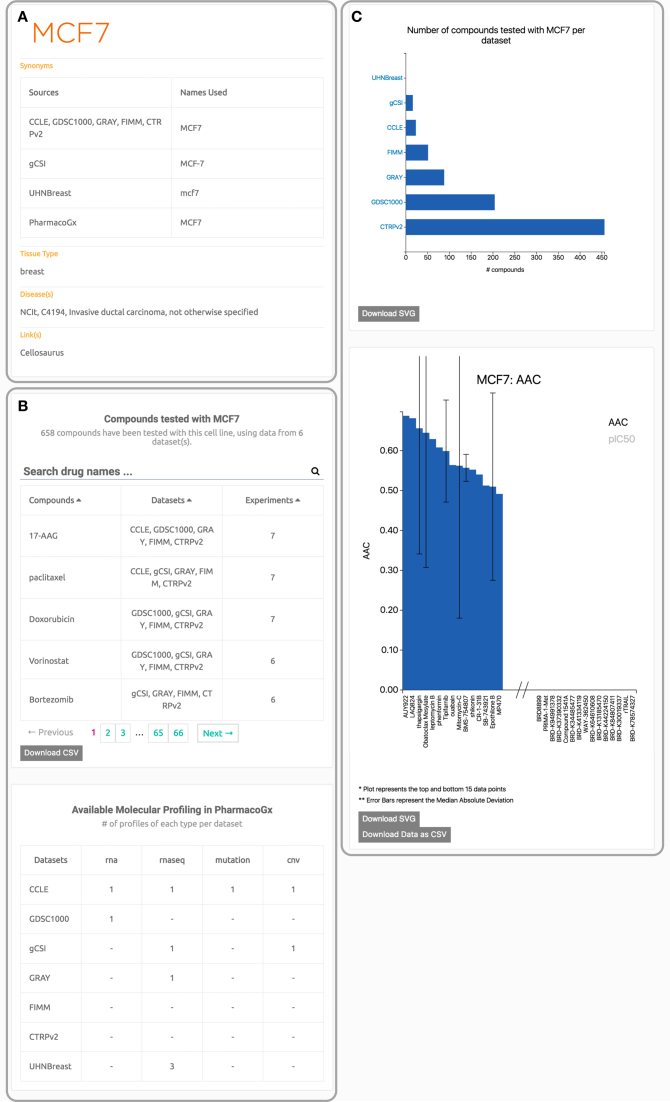
Profile pages
If a search query for a single entity is entered into the search bar, or if an entity is selected in the explore page, the user is redirected to a profile page. This page is designed to provide the user with a comprehensive view of all the data available for the entity of interest (Figure 3). Textual information is consistently positioned on the left side, and visualizations of the data are on the right. Each page contains a card in the top left corner describing the entity selected, and any relevant metadata available for this entity. For example, a cell line card would contain the relevant annotations and a description of the available molecular and pharmacological data collected for each dataset, the tissue and disease type for a cell line (Figure 3A). The card will also contain links to any relevant external databases, such as GeneCards for targets, Pubchem for compounds, and Cellosaurus for cell lines. Searchable tables list all the dose-response experiments in PharmacoDB pertaining to the entity being profiled (Figure 3B). Elements in these tables tables are fully linked to allow users to navigate between profile pages in PharmacoDB, or by clicking on the experiment count for a compound and cell line pair, redirected directly to page displaying the drug testing experiment(s). The right-hand side of the profile pages displays plots with summary statistics about the entity (Figure 3C). For a cell line, a waterfall plot reports the 15 compounds with the lowest and highest efficacy or potency. These plots allow finding the most effective drug for a cell line of interest, or finding cell lines which are abnormally sensitive for a drug of interest, across all the datasets included in the database.
Figure 3.
An example profile page from PharmacoDB for a cell line. The page is organized so that the left column contains textual information, and the right column contains plots. Panel (A) is the information card for the cell line (MCF7). Panel (B) contains tables listing the available data profiles for this cell. Panel (C) contains summary plots about the drug screening performed in each dataset and the waterfall plots of the cell response to treatment with compounds.
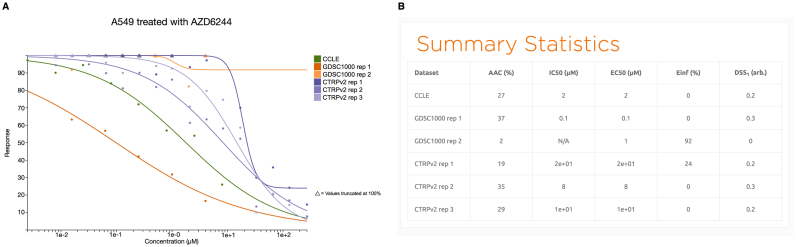
Drug dose-response curves
Searching for a cell line-drug pair or selecting a cell line with a drug through the explore page will redirect to a page displaying the dose-response data found across all datasets. The page includes a plot of the measured viability values and a Hill Slope curve fit to the measured data (Figure 4A; Supplementary Methods), followed by a table of summary statistics commonly used to summarize the (in)sensitivity of the cell line to the given compound (Figure 4B). Each curve plotted on the graph can be hidden and shown by clicking on its entry in the legend. We used PharmacoGx (19) to normalize and reprocess all cell viability data with a uniform pipeline to remove any biases between datasets introduced by computational aspects such as choice of Hill Slope model, curve fitting algorithms, or inconsistent calculations of summary statistics between studies (Supplementary Figure S1; Supplementary Methods). Given the lack of consensus regarding the best way to summarize drug dose-response curve, we computed a compendium of summary metrics for the response of the cell line to the treatment with the compound, including the common IC50 (dose of 50% inhibition of cell viability), EC50 (dose at which 50% of the maximum response is observed), Area Above Curve (AAC), Einf (maximum theoretical inhibition), and the recent drug sensitivity score (DSS) (32). Hovering over a value in the summary measures table will display on the plot a visualization of the procedure used to calculate the summary statistic. As there is no consensus as to the optimal metric for summarizing the information contained in the dose response curve (33). The IC50 and EC50 metrics focus on the potency of the compound, the Emax on the efficacy, and the AAC and DSS integrate both potency and efficacy. These metrics are presented together on PharmacoDB, and a visualization of method of calculating them is displayed directly on the drug dose-response curve to aid in making an informed decision of the correct measure to use for a given experiment or specific biological question of interest.
Figure 4.
An example of drug dose response curve plot with (A) the A549 lung cancer cell line treated with the MEK inhibitor AZD6244, and (B) the corresponding table of summary statistics. IC50: dose of 50% inhibition of cell viability; EC50: dose at which 50% of the maximum response is observed; AAC: area above curve; Einf: maximum theoretical inhibition; DSS: drug sensitivity score.
Batch query
For queries involving multiple cell lines and compounds, users can access the drug sensitivity data via the Batch Query page. While the search interface is limited to the retrieval of drug sensitivity data for one compound / cell-line pair at a time, the batch query interface allows users to quickly cut and paste their list of cell lines and compounds of interest. After submission, a spreadsheet containing all the summary metrics for the drug dose-response curves included in PharmacoDB will be available for download.
PharmacoGx
In addition to cell line viability screens, the cancer pharmacogenomic datasets included in PharmacoDB include extensive molecular profiling. We recently released the PharmacoGx package (19) to facilitate the analysis of the relationships between the pharmacological and molecular data for the purposes of biomarker discovery (18) and drug repurposing (34). The reprocessing of pharmacological data and the extensive curation of identifiers done for PharmacoDB has been fully integrated into PharmacoSet (PSet) R objects released with the PharmacoGx platform. While PharmacoDB does not contain molecular data, a PSet object has been created and linked to from each dataset profile page (Supplementary Figure S1A). To facilitate finding molecular data for a specific cell line, a table describing the availability of molecular profiles in PharmacoGx is available at the bottom of each cell line profile page (Figure 3). Furthermore, each compound profile page and gene profile page in PharmacoDB includes a table of univariate associations between the molecular features of the cell lines included in the database and their response to compounds they were tested with (Supplementary Figure S5). These associations were computed using the drugSensitivitySig function in PharmacoGx, described in more detail in Supplementary Methods. This link between PharmacoDB and PharmacoGx enables bioinformaticians to use the web-application as an entry point for their pharmacogenomic analysis, and allows them to leverage our extensive curation. As an example, one can quickly use PharmacoDB to verify that high expression of the gene ERBB2 predisposes cell lines to be sensitive to treatment with lapatinib. Searching ERBB2 brings up the gene profile page, where lapatinib is listed as a top associated drug with moderate effect size and very high significance. This association was found in CCLE, which suggests to researchers interested in investigating this association further that CCLE would be a good starting point for their analysis. Downloading the CCLE PSet as instructed on the link from the CCLE profile page, one can delve deeper using PharmacoGx, for instance to repeat this analysis per tissue type. Example code for such an analysis is provided in Supplementary Methods, and reveals as expected that the association is strongest in breast (Standardized Coefficient: 0.75, p-value: 2.7 × 10−06), and soft tissue breast (standardized coefficient: 0.85, P-value: 2.3 × 10−04) and weak in hematopoietic/lymphatic and bone (standardized coefficient: –0.07, P-value: 0.92 and standardized coefficient: 0.04, P-value: 0.56 respectively).
USER ACCESS TO DATA, CODE AND FEEDBACK
Programmatic data access
PharmacoDB exposes its data through an Application Programming Interface (API), enabling users to programmatically interact with the application. The API is RESTful (Representational State Transfer), meaning that all application resources are made available using a predefined set of stateless operations, in this case being HTTP verbs such as GET, POST, DELETE. No authentication keys, or tokens, are currently needed in order to access the API. The API has been implemented using the Go programming language, and Gin HTTP web framework has been used for routing. All queries are made using HTTP GET requests, and all results are returned in JSON format by default. All the data in PharmacoDB are publicly available via the API. Additionally, a dump of the SQL database is available for download from the front page and R objects for all the pharmacogenomic datasets are available via the PharmacoGx R/Bioconductor package (19).
Code and documentation
The PharmacoDB code is open-source and publicly available through the PharmacoDB GitHub repository (github.com/bhklab/PharmacoDB) under the GPLv3 license. The documentation is available in the web-application as video and textual descriptions of all the entities and search queries in PharmacoDB. These include descriptions of the dataset, tissue, cell line, genes, and drug/compound pages. Tutorials on how to perform more complex queries, such as displaying a drug dose-response curve and intersecting datasets, are also described in detail in the Documentation page of PharmacoDB.
Feedback
Our web-application provides an easily accessible, optionally anonymous contact mechanism for providing feedback on all aspects of PharmacoDB. Users can suggest corrections to annotations by clicking on the feedback icon accessible on the left of every profile page. The fields in the ‘Contact Us’ page are then prefilled with data relevant to the annotation in question. The GitHub API is then used to automatically file user suggestions and feedback as issues in the GitHub Issue Tracker at our repository (github.com/bhklab/PharmacoDB). This allows for full transparency regarding the reliability of data in the database and enables the community to fully assess and correct any missing information.
SUMMARY AND FUTURE DIRECTIONS
PharmacoDB is the first database providing a comprehensive resource to search and explore the largest pharmacogenomic studies released to date. By combining rigorous curation of identifiers across the published pharmacogenomic datasets with comprehensive search and visualizations of the pharmacological data, PharmacoDB allows researchers to quickly access the data available to answer their biological questions of interest. It provides an interface to query for specific drug dose-response curves, and easily find the largest possible intersection between datasets.
As current pharmacogenomic datasets continue to expand and new ones are published, the number of cell lines screened with compounds will increase, opening new avenues of research for meta-analysis in biomarker discovery and other applications. In this setting, PharmacoDB will provide a unique resource where researchers can quickly mine the large amount of data generated by these high-throughput drug screening studies. Moreover, given the recent activity in the pharmacogenomic field, new statistical approaches are being developed to better model and summarize drug dose-response curves. Recently, Hafner et al. published the growth rate inhibition 50 (GR50) metric to robustly quantify drug response by accounting for the different proliferation rate of each cancer cell lines (35), and showed an increase in consistency across datasets (36). Although this method and others may require data that are not always available for all datasets (e.g., proliferation rate of each cell lines for GR50) we are committed to implement them to provide users with the opportunity to select the most relevant readout for their specific application. In addition to datasets measuring cell viability, we also plan to update PharmacoDB with pharmacogenomic datasets reporting the transcriptional changes due to chemical perturbation, such as the Connectivity Map (37) and the L1000 (Subramanian et al., BiorXiv 2017) datasets. The combination of drug sensitivity and perturbation data would allow users to study deeper the relationship between the molecular state of cancer/normal cell lines and their response to compound perturbations (34). Other datasets assessing the toxic effect of chemical perturbations in hepatocytes and kidney cell lines (38–40) will also be integrated to extend the scope of PharmacoDB beyond cancer. The flexibility of PharmacoDB will enable continuous update of the pharmacogenomic datasets, and facilitate the analysis of these valuable data by the scientific community.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the investigators of the Genomics of Drug Sensitivity in Cancer (GDSC), the Cancer Cell Line Encyclopedia (CCLE), Genentech (gCSI), the Institute for Molecular Medicine Finland (FIMM), Drs Joe W. Gray, Benjamin G. Neel and David W. Cescon who have made their valuable pharmacogenomic data available to the scientific community. The authors also thank Heather Selby for her help curating the cell line identifiers, and Aimos Bairoch for developing Cellosaurus and helping us obtain detailed annotations for our collection of cell lines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Cancer Society Research Institute and the Canadian Institutes for Health Research (to P.S.); Terry Fox Research Institute and the Cancer Research Society (to Z.S.); Canadian Cancer Society Research Institute (to P.S.); Terry Fox Research Institute (to W. Ba-alawi); Gattuso Slaight Personalized Cancer Medicine Fund at Princess Margaret Cancer Centre, the Canadian Institute of Health Research and Natural Sciences and Engineering Research Council, the Ontario Institute for Cancer Research, and the Stand Up To Cancer Canada (to B. Haibe-Kains). Funding for open access charge: Princess Margaret Cancer Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Global Burden of Disease Cancer Collaboration Fitzmaurice C., Allen C., Barber R.M., Barregard L., Bhutta Z.A., Brenner H., Dicker D.J., Chimed-Orchir O., Dandona R. et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017; 3:524–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoadley K.A., Yau C., Wolf D.M., Cherniack A.D., Tamborero D., Ng S., Leiserson M.D.M., Niu B., McLellan M.D., Uzunangelov V. et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014; 158:929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ludwig J.A., Weinstein J.N.. Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer. 2005; 5:845–856. [DOI] [PubMed] [Google Scholar]
- 4. Garraway L.A., Verweij J., Ballman K.V.. Precision oncology: an overview. J. Clin. Oncol. 2013; 31:1803–1805. [DOI] [PubMed] [Google Scholar]
- 5. Sharma S.V., Haber D.A., Settleman J.. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat. Rev. Cancer. 2010; 10:241–253. [DOI] [PubMed] [Google Scholar]
- 6. Macarron R., Banks M.N., Bojanic D., Burns D.J., Cirovic D.A., Garyantes T., Green D.V.S., Hertzberg R.P., Janzen W.P., Paslay J.W. et al. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 2011; 10:188–195. [DOI] [PubMed] [Google Scholar]
- 7. Garnett M.J., Edelman E.J., Heidorn S.J., Greenman C.D., Dastur A., Lau K.W., Greninger P., Thompson I.R., Luo X., Soares J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012; 483:570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daemen A., Griffith O.L., Heiser L.M., Wang N.J., Enache O.M., Sanborn Z., Pepin F., Durinck S., Korkola J.E., Griffith M. et al. Modeling precision treatment of breast cancer. Genome Biol. 2013; 14:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H., Degenhardt J., Mayba O., Gnad F., Liu J. et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015; 33:306–312. [DOI] [PubMed] [Google Scholar]
- 11. Haverty P.M., Lin E., Tan J., Yu Y., Lam B., Lianoglou S., Neve R.M., Martin S., Settleman J., Yauch R.L. et al. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature. 2016; 533:333–337. [DOI] [PubMed] [Google Scholar]
- 12. Mpindi J.P., Yadav B., Östling P., Gautam P., Malani D., Murumägi A., Hirasawa A., Kangaspeska S., Wennerberg K., Kallioniemi O. et al. Consistency in drug response profiling. Nature. 2016; 540:E5–E6. [DOI] [PubMed] [Google Scholar]
- 13. Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Gonçalves E., Barthorpe S., Lightfoot H. et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016; doi:10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hatzis C., Bedard P.L., Juul Birkbak N., Beck A.H., Aerts H.J.W.L., Stern D.F., Shi L., Clarke R., Quackenbush J., Haibe-Kains B.. Enhancing reproducibility in cancer drug screening: how do we move forward?. Cancer Res. 2014; doi:10.1158/0008-5472.CAN-14-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haverty P.M., Lin E., Tan J., Yu Y., Lam B., Lianoglou S., Neve R.M., Martin S., Settleman J., Yauch R.L. et al. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature. 2016; 533:333–337. [DOI] [PubMed] [Google Scholar]
- 16. Cancer Cell Line Encyclopedia Consortium and Genomics of Drug Sensitivity in Cancer Consortium Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015; 528:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Safikhani Z., El-Hachem N., Quevedo R., Smirnov P., Goldenberg A., Juul Birkbak N., Mason C., Hatzis C., Shi L., Aerts H.J. et al. Assessment of pharmacogenomic agreement. F1000Res. 2016; 5:825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Safikhani Z., Smirnov P., Freeman M., El-Hachem N., She A., Rene Q., Goldenberg A., Juul-Birkbak N., Hatzis C., Shi L. et al. Revisiting inconsistency in large pharmacogenomic studies. F1000Res. 2016; 5:2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smirnov P., Safikhani Z., El-Hachem N., Wang D., She A., Olsen C., Freeman M., Selby H., Gendoo D.M., Grossman P. et al. PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics. 2016; doi:10.1093/bioinformatics/btv723. [DOI] [PubMed] [Google Scholar]
- 20. Seashore-Ludlow B., Rees M.G., Cheah J.H., Cokol M., Price E.V., Coletti M.E., Jones V., Bodycombe N.E., Soule C.K., Gould J. et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015; doi:10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Basu A., Bodycombe N.E., Cheah J.H., Price E.V., Liu K., Schaefer G.I., Ebright R.Y., Stewart M.L., Ito D., Wang S. et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013; 154:1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heiser L.M., Sadanandam A., Kuo W.-L., Benz S.C., Goldstein T.C., Ng S., Gibb W.J., Wang N.J., Ziyad S., Tong F. et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2011; 109:2724–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bairoch A. ExPASy - Cellosaurus. Cellosaurus. 2015; http://web.expasy.org/cellosaurus/. [Google Scholar]
- 24. Corsello S.M., Bittker J.A., Liu Z., Gould J., McCarren P., Hirschman J.E., Johnston S.E., Vrcic A., Wong B., Khan M. et al. The drug repurposing hub: a next-generation drug library and information resource. Nat. Med. 2017; 23:405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V. et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014; 42:D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M., Krüger F.A., Light Y., Mak L., McGlinchey S. et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014; 42:D1083–D1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Safran M., Chalifa-Caspi V., Shmueli O., Olender T., Lapidot M., Rosen N., Shmoish M., Peter Y., Glusman G., Feldmesser E. et al. Human gene-centric databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003; 31:142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rebhan M., Chalifa-Caspi V., Prilusky J., Lancet D.. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998; 14:656–664. [DOI] [PubMed] [Google Scholar]
- 29. Whirl-Carrillo M., McDonagh E.M., Hebert J.M., Gong L., Sangkuhl K., Thorn C.F., Altman R.B., Klein T.E.. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012; 92:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaplun A., Hogan J.D., Schacherer F., Peter A.P., Krishna S., Braun B.R., Nambudiry R., Nitu M.G., Mallelwar R., Albayrak A.. PGMD: a comprehensive manually curated pharmacogenomic database. Pharmacogenomics J. 2016; 16:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dalabira E., Viennas E., Daki E., Komianou A., Bartsakoulia M., Poulas K., Katsila T., Tzimas G., Patrinos G.P.. DruGeVar: an online resource triangulating drugs with genes and genomic biomarkers for clinical pharmacogenomics. Public Health Genomics. 2014; 17:265–271. [DOI] [PubMed] [Google Scholar]
- 32. Yadav B., Pemovska T., Szwajda A., Kulesskiy E., Kontro M., Karjalainen R., Majumder M.M., Malani D., Murumägi A., Knowles J. et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci. Rep. 2014; 4:5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fallahi-Sichani M., Honarnejad S., Heiser L.M., Gray J.W., Sorger P.K.. Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat. Chem. Biol. 2013; 9:708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. El-Hachem N., Gendoo D.M.A., Ghoraie L.S., Safikhani Z., Smirnov P., Chung C., Deng K., Fang A., Birkwood E., Ho C. et al. Integrative Cancer Pharmacogenomics to Infer Large-Scale Drug Taxonomy. Cancer Res. 2017; 77:3057–3069. [DOI] [PubMed] [Google Scholar]
- 35. Hafner M., Niepel M., Chung M., Sorger P.K.. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat. Methods. 2016; 6:521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hafner M., Niepel M., Sorger P.K.. Alternative drug sensitivity metrics improve preclinical cancer pharmacogenomics. Nat. Biotechnol. 2017; 35:500–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.-P., Subramanian A., Ross K.N. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006; 313:1929–1935. [DOI] [PubMed] [Google Scholar]
- 38. El-Hachem N., Grossmann P., Blanchet-Cohen A., Bateman A.R., Bouchard N., Archambault J., Aerts H.J.W.L., Haibe-Kains B.. Characterization of Conserved Toxicogenomic Responses in Chemically Exposed Hepatocytes across Species and Platforms. Environ. Health Perspect. 2016; 124:313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Igarashi Y., Nakatsu N., Yamashita T., Ono A., Ohno Y., Urushidani T., Yamada H.. Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res. 2015; 43:D921–D927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ganter B., Snyder R.D., Halbert D.N., Lee M.D.. Toxicogenomics in drug discovery and development: mechanistic analysis of compound/class-dependent effects using the DrugMatrix database. Pharmacogenomics. 2006; 7:1025–1044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.