Abstract
Carbohydrate-active enzyme (CAZymes) are not only the most important enzymes for bioenergy and agricultural industries, but also very important for human health, in that human gut microbiota encode hundreds of CAZyme genes in their genomes for degrading various dietary and host carbohydrates. We have built an online database dbCAN-seq (http://cys.bios.niu.edu/dbCAN_seq) to provide pre-computed CAZyme sequence and annotation data for 5,349 bacterial genomes. Compared to the other CAZyme resources, dbCAN-seq has the following new features: (i) a convenient download page to allow batch download of all the sequence and annotation data; (ii) an annotation page for every CAZyme to provide the most comprehensive annotation data; (iii) a metadata page to organize the bacterial genomes according to species metadata such as disease, habitat, oxygen requirement, temperature, metabolism; (iv) a very fast tool to identify physically linked CAZyme gene clusters (CGCs) and (v) a powerful search function to allow fast and efficient data query. With these unique utilities, dbCAN-seq will become a valuable web resource for CAZyme research, with a focus complementary to dbCAN (automated CAZyme annotation server) and CAZy (CAZyme family classification and reference database).
INTRODUCTION
Significance of CAZymes
CAZymes (carbohydrate-active enzymes) are involved in complex carbohydrate metabolism. They are responsible for the synthesis (through glycosyltransferases [GTs]), degradation (glycoside hydrolases [GHs], polysaccharide lyases [PLs], carbohydrate esterases [CEs], and enzymes for the auxiliary activities [AAs]) and recognition (carbohydrate-binding module [CBM]) of all the carbohydrates on Earth (1). CAZymes are found in all living organisms (typically 1–3% of the gene content) and particularly abundant (>3% of the gene content) in plants and plant degrading/saprophytic/pathogenic microbes (2–4) (Table 1). The reason is that a large number of CAZymes are needed to build (in plants) and degrade (in microbes) the complex carbohydrates of plant cell walls. Particularly, microbes living in the animal guts encode the highest percentage of CAZymes degrading various diet-derived carbohydrates and host carbohydrates (Table 1), and changing the dietary carbohydrates has a major impact on the microbiota structure in the human guts and further influences the human health (5,6).
Table 1. CAZymes are most abundant in plants and plant-associated microbes.
| Organism | # of CAZymes | % of gene content |
|---|---|---|
| Arabidopsis thaliana (plant) | 1,134 | 4.1% |
| Aspergillus oryzae (plant-associated fungus) | 506 | 3.9% |
| Bacteroides thetaiotaomicron (human gut bacterium) | 391 | 8.2% |
| Escherichia coli MG1655 (laboratory bacterium) | 106 | 2.6% |
| Homo sapiens (human) | 340 | 1.7% |
Similar online resources
The CAZy database (1) started to collect experimentally characterized CAZyme proteins from literature and classify them into protein families based on sequence similarity since 1990s. It then populated each family by including homologs of the characterized seed proteins from GenBank, UniProt and PDB databases. Although CAZy is most well known as a CAZyme classification database, it also provides an HTML page for each CAZyme family, presenting that family's known enzymatic activities (i.e. EC numbers), as well as information about its member proteins, e.g. protein IDs, names, and species. However, it does not provide a way to download CAZyme sequence and annotation data, nor a sequence search service to automatically annotate given genomes for CAZymes.
In 2012, we developed a web server, dbCAN (7), together with two other tools CAT (8) and Hotpep (9) developed by others, to allow automated CAZyme annotation for newly sequenced genomes/metagenomes. Updated and well maintained every year, dbCAN has become the most popular automated CAZyme annotation server.
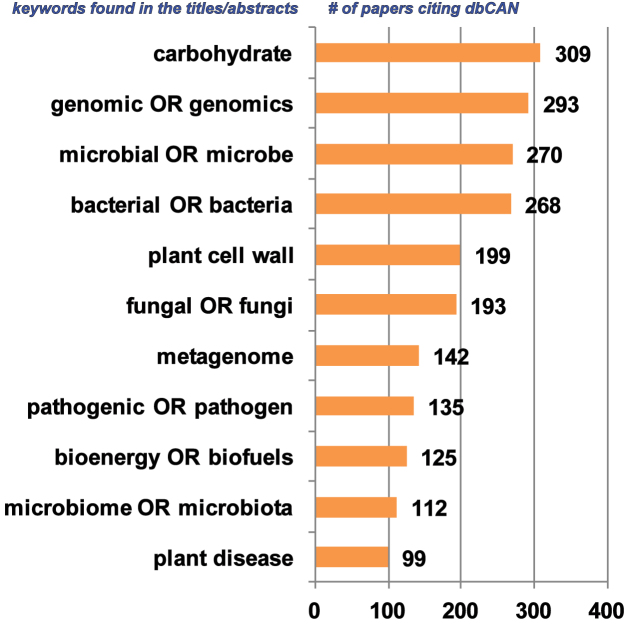
Since 2012, we were frequently contacted by dbCAN users about the availability of pre-computed CAZyme sequence and annotation data for sequenced/published genomes and metagenomes. In 2014, we developed a pre-computed CAZyme database for fully sequenced plant and algal genomes named PlantCAZyme (10). Furthermore, a survey of publications citing dbCAN using Google Scholar found that dbCAN has been most frequently used to annotate microbial genomes and metagenomes (Figure 1).
Figure 1.
dbCAN has been cited in various research fields.
Motivations
Apparently, other than plant biologists, there are substantially more researchers from a variety of research fields (Figure 1), such as genomics, carbohydrate, bioenergy, plant disease, food security, microbial ecology, and human microbiome, who are more interested in microbial CAZymes. Hence, with tens of thousands of bacterial genomes available, we built the dbCAN-seq database (http://cys.bios.niu.edu/dbCAN_seq), aiming to provide a one-stop online database with the most comprehensive pre-computed microbial CAZyme sequence and annotation data.
DATABASE CONTENT
CAZyme identification and annotation
We scanned 5,349 completely assembled bacterial genomes (proteomes) of the RefSeq database to identify CAZymes. These 5,349 genomes are from 4,056 unique taxonomic IDs and 2,192 unique species (Supplementary Table S1), meaning that one species can have different taxonomic IDs and strains, and one taxonomic ID can also have different genome assemblies of different GCF (RefSeq assembly) IDs.
Specifically, we followed the stand-alone dbCAN annotation pipeline (http://csbl.bmb.uga.edu/dbCAN/download/readme.txt), to keep dbCAN hits with E-value < 1e–5 and coverage > 0.3. We found in total 572,269 CAZyme homologs in 5,329 bacterial genomes. If using a more stringent threshold (E-value < 1e–18 and coverage > 0.35), 330,307 homologs of 5,288 bacterial genomes will remain. According to a benchmark analysis similar to (10), this stringent threshold resulted in an F-measure = 2 × (Recall × Precision)/(Recall + Precision) at 90.1% for Escherichia coli MG1655, 93.1% for Clostridium thermocellum ATCC 27405, and 89.9% for Anaerocellum thermophilum DSM 6725 (Supplementary Tables S2–S4). In our website, we still provided the data for the 572,269 CAZyme homologs together with the E-value and coverage information in case the users want to filter the homologs for more reliable CAZymes.
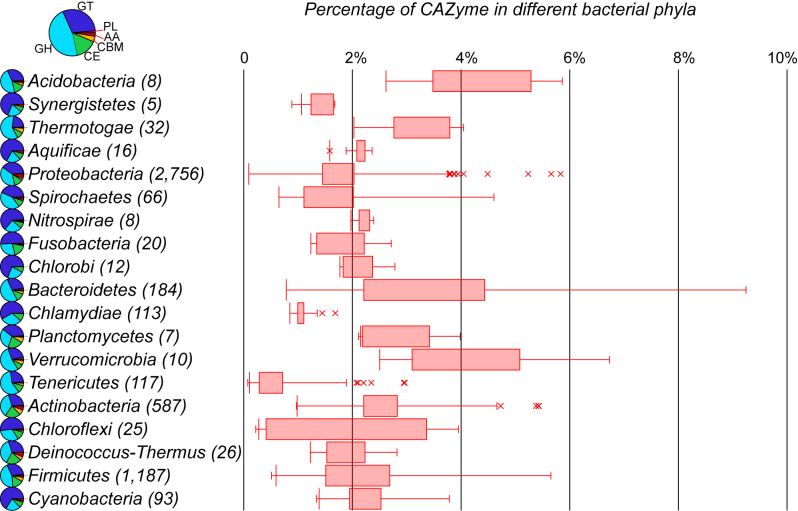
Figure 2 shows a box plot of the percentage of CAZymes (the number of CAZymes divided by the total number of proteins in a proteome). It is clear that Acidobacteria (8 genomes), Verrucomicrobia (10 genomes), Bacteroidetes (184 genomes), and Thermotogae (32 genomes) are among the top bacterial phyla having the highest percentages, which tend to also have higher fractions of carbohydrate-degrading enzymes (GH+CE+PL in the pie charts of Figure 2). This is not surprising as bacteria of these phyla are well known for their ability to degrade various complex carbohydrates in plant/algal material-rich environments such as animal guts, marine sediment, and soils (11–14).
Figure 2.
CAZymes in different phyla. Pie charts (left): the relative fraction of different CAZyme classes, which include GTs (glycosyltransferases), GHs (glycoside hydrolases), PLs (polysaccharide lyases), CEs (carbohydrate esterases), AAs (enzymes of the auxiliary activities), and CBMs (carbohydrate-binding modules). Box plots (right): the percentage of CAZymes in different bacterial phyla. The number in the parentheses is the number of genomes.
After extracted the CAZyme protein sequences, we further performed extensive functional annotation for each CAZyme, including predictions of enzyme EC numbers using E2P2 (15), dbCAN signature domains (7), CDD functional domains (16), sequence homologs in databases CAZy (1), PDB (17), Swiss-Prot (18), as well as predictions of signal peptides using SignalP (19), lipoproteins using LipoP (20), and transmembrane domains using TMHMM (21), and lastly, protein secondary structures using PSSpred (22). We also collected data from the RefSeq database and calculated the basic information for each CAZyme, e.g. protein length, molecular weight, isoelectric point, genomic location and genomic context (gene neighborhood).
CAZyme gene clusters (CGCs) identification
Human gut-associated Bacteroidetes genomes were found to encode dozens of polysaccharide utilization loci (PULs), defined as physically linked genes specializing in the breakdown of various dietary fiber carbohydrates (23–26). A recent computational PUL prediction was performed in 67 Bacteroidetes genomes using SusC (sugar transporter) and SusD (glycan binding protein) genes as signature genes and CAZymes as accessory genes (27).
The finding of PULs strongly suggests that CAZymes often work together with each other and with transcription factors (TFs) and transporters (TCs) to synergistically degrade/synthesize various highly complex carbohydrates. It is very likely that in nature there exist numerous PUL-like gene clusters in more bacteria in addition to Bacteroidetes, which may contain other transporters than SusC/D. Identifying CAZyme-containing gene clusters and presenting them on the web will be a valuable resource to the carbohydrate research community, especially to researchers interested in identifying and characterizing new PULs using wet-lab approaches.
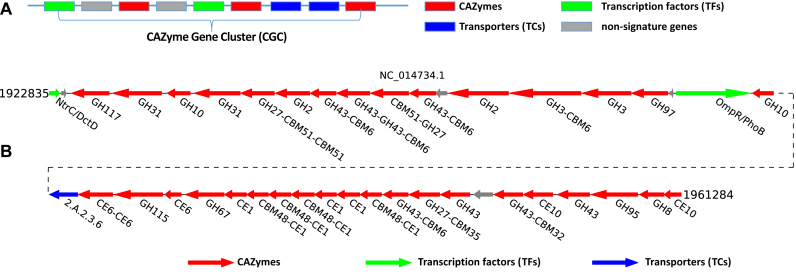
We defined a more general term CAZyme gene clusters (CGCs) to be physically linked gene clusters that must contain three different classes of signature genes: (i) CAZymes, (ii) TFs and (iii) TCs. Between two adjacent signature genes, a small number of non-signature genes are also allowed (Figure 3A). CAZyme identification was already described above. For the other two classes of signature genes, we searched against TCDB for TCs (28), and searched against CollectTF (29), DBTBS (30) and RegulonDB (31) for TFs, by using DIAMOND (32) with an E-value cutoff < 1e–10.
Figure 3.
CAZyme gene cluster (CGC) definition and example. (A) Definition of CGC: One CGC must contain at least one CAZyme (red). Two other signature gene classes could also be present: TF (green) and TC (blue) genes. A small number of non-signature genes (gray) can be inserted between two neighboring signature genes. (B) An example CGC from Paludibacter propionicigenes WB4, which has 42 genes in one cluster including 35 CAZymes (red), 1 TC (blue), 2 TFs (green) and 4 other genes (gray). The CAZyme gene labels are based on CAZyme domain assignment. The TC gene labels are based on the best hit in TC-DB. The TF gene labels are based on the best hit in a few TF databases (see main text).
We then developed a python program called CGC-Finder to scan the 5,329 bacterial genomes for CGCs. The inputs to this program include a signature gene annotation file and a gene location file. Two important parameters are needed: (i) the distance threshold (range between 0 and 10) to allow a certain number of non-signature genes inserted between two adjacent signature genes; (ii) the signature gene classes (CAZyme+TF+TC, CAZyme+TC, or CAZyme+TF) that must be present in the CGC. These parameters allow the users to define a CGC more loosely or more stringently at their will: e.g. all three signature gene classes have to be present and a minimum number of non-signature genes are allowed in the CGC.
Using a stringent parameter setting (distance < 2 and CAZyme+TF+TC), we found in total 26,397 CGCs in 4,077 genomes (Supplementary Table S5). The largest CGC is Paludibacter propionicigenes WB4’s NC_014734.1-CGC2 (Bacteroidetes phylum) containing in total 42 genes (Figure 3B). Among the 4,077 genomes (Supplementary Table S6), Streptomyces bingchenggensis BCW-1 (Actinobacteria phylum) has the largest number (34) of CGCs, the largest number (234) of signature genes, and the largest number (291) of total genes in the CGCs. At the same time, it also has one of the largest genomes (11.8Mb). On the other hand, Bacteroides cellulosilyticus (Bacteroidetes phylum) with a genome size at 7.1 Mb has the largest number (92) of CAZyme genes located in the CGCs, and the highest percentage (11.7%) of CAZymes overall.
To remove the effect of genome size, we further calculated the percentage of genes located in CGCs (the number of genes in CGCs divided by the total number of genes in the genome), and the percentage of CAZymes located in CGCs (the number of CAZymes in CGCs divided by the total number of CAZymes in the genome) (Supplementary Table S6). We found that Bifidobacterium scardovii JCM 12489 (Actinobacteria phylum) has the highest percentage of CAZymes (35.1%) located in CGCs and the highest percentage of total genes (8.9%) located in CGCs, suggesting CAZymes of this genome have a strong tendency to be clustered into CGCs.
It should be emphasized that our CGCs are defined differently than the classic PULs in the literature. CGC is a broader term than PUL, and is created to study which CAZymes are physically clustered with which other CAZymes, TFs and TCs. Most published PULs were defined in some individual genomes or in some particular group of bacteria (e.g. Bacteroidetes), with a strong bias in favor of SusC/D (TCs) and without requirement of TFs, also often followed by experimental verification. Therefore, it is not meaningful to compare published PULs with CGCs identified here in a much larger scale.
Species metadata collection
We have extracted the species sample information of the 5,349 genomes using the JGI IMG database's metadata table (33) by matching the NCBI BioSample IDs. We showed in Supplementary Table S7 that: (i) 459 genomes have Disease information; (ii) 1,245 have Ecosystem info; (iii) 1,165 have Ecosystem Category info; (iv) 1,124 have Ecosystem Subtype info; (v) 1,143 have Ecosystem Type info; (vi) 1,200 have Habitat info; (vii) 321 have Metabolism info; (viii) 1,089 have Motility info; (ix) 1,138 have Oxygen Requirement info; (x) 684 have Phenotype info; (xi) 242 have Sample Body Site info; (xii) 1,111 have Specific Ecosystem info; (xiii) 1,188 have Temperature Range info. All these metadata was incorporated into the dbCAN-seq database and used to classify the genomes into different metadata groups.
WEB DESIGN
The dbCAN-seq website is powered by MySQL+PHP+JavaScript+Sphinx. The following features are unique to dbCAN-seq and not available at other CAZyme websites:
Download page
The download page has a searchable table with all the 5,329 genomes. Each row of the table corresponds to a genome with a download link to a compressed tarball file. The tarball contains a FASTA sequence file of all the CAZymes in the genome, and a tab-separated file with all the annotation and location data. There is also a link to download data of all the genomes.
CGC page
The CGC page is designed as a tool page. When a user opens the ‘CAZyme Gene Cluster’ page, a table will be seen with all the 5,329 genomes. The table actually has 7,841 rows as one genome can have multiple RefSeq IDs (chromosomes and plasmids). One can click on the genome to open a new page, where two parameters need to be set: (i) distance and (ii) signature gene classes, which were already described above. After hitting on the ‘Calculate’ button, the CGC-Finder python program will be called to identify CGCs in the genome. The program runs very fast: processing one genome takes a few milliseconds, and processing all the 5,329 genomes together takes < 1 min.
The CGC-Finder result will be printed as a table below the parameter selection section. Each row in the table is one CGC with different statistics about the CGC, such as the numbers of the three signature genes and the number of all genes (including non-signature ones). Clicking on the CGC_no will open a separate page showing: (i) the graphical representation of the genomic location of the CGC in a Jbrowser; (ii) at the bottom the detailed information about all genes in the CGC as a table, such as the genomic location, the functional description, and if signature gene and evidence. All the data tables can be downloaded by clicking on a download link above the table.
Metadata page
The metadata page has a pull-down menu, where users can select one from 16 different metadata types. Under the pull-down menu, a bar graph is shown with each bar representing one group of the selected metadata type. For example, one particular disease Tuberculosis is associated with 16 genomes, and the height of the bar is 16. Clicking on the bar will open a new page with another bar graph, now each bar representing one genome and the height representing the number of CAZymes in that genome. This design allows users to quickly browse a variety of metadata types, the genomes associated with one particular group of metadata, and the CAZymes in the genome.
Protein annotation page
The protein annotation page collects all kinds of annotation about a CAZyme and presents them in 13 sections: basic information, genomic context (graphical JBrowser window), full-length sequence (protein and nucleotide CDS sequence), enzyme annotation, CAZyme signature domains, CDD domains, CAZy hits, PDB hits, Swiss-Prot hits, signal peptide prediction, transmembrane prediction, protein secondary structure prediction, and lipoprotein prediction. All the 572 269 CAZymes have a protein page, which is dynamically generated by a PHP script querying different MySQL tables. From this page, users can quickly browse/obtain the most comprehensive pre-computed CAZyme sequence and annotation data.
Search function
A search box is shown at the top part of all pages. The search function is built upon the powerful search engine Sphinx, which has been programmatically configured to allow very fast index-organized table search (average response time: 300 ms) and highly efficient pagination. It implements a Google-like search supporting both exact and fuzzy query, and users can input a keyword to search 12 different data types. These data types can be largely classified into three groups: (i) basic information about CAZymes: such as species name, CAZyme domain, protein ID, GCF ID, taxonomy ID; (ii) CAZyme annotation data: such as PDB hits, Swiss-Prot hits, CAZy hits, CDD hits, E2P2 predicted enzyme reaction and EC number; (iii) CAZyme genomic context.
For data types in (ii), it is to search for CAZymes sharing sequence similarity to Swiss-Prot, PDB, and CAZy proteins. For example, one can type in a PDB ID (e.g. 1LZL_A) and choose a sequence identity value (e.g, 50%). The search will return a list of CAZymes sharing similarity with the queried PDB protein with identity larger than the given value: http://cys.bios.niu.edu/dbCAN_seq/search.php?sim=50&signum=5&search_type=6&search_txt=1LZL_A. This is very useful to answer questions like, what proteins in dbCAN-seq have a high sequence similarity to some experimentally characterized CAZymes?
As for (iii), it is a very useful tool to search the gene neighborhood of a query CAZyme. For example, users can type in a CAZyme protein ID (e.g. WP_007212487.1), and select how many upstream and downstream genes of the query gene they want to explore (e.g. 5). The search will return a table with 11 genes with the query gene being the sixth in the table: http://cys.bios.niu.edu/dbCAN_seq/search.php?sim=20&signum=5&search_type=12&search_txt=WP_007212487.1. If any of the 11 genes are CGC signature genes (i.e. CAZyme, TF or TC), they will be highlighted with colors. This is a novel tool to answer questions like, is my CAZyme located close to any other CAZyme genes or TF or TC genes, or is my CAZyme potentially located in any CGCs?
CONCLUSIONS
Compared to other CAZyme web resources, dbCAN-seq has the following unique features:
It provides a convenient download page for users to batch download all the pre-computed CAZyme sequence and annotation data for 5,329 fully sequenced bacterial genomes;
It provides the most comprehensive annotation data for computationally identified CAZymes;
It provides a metadata page to organize the bacterial genomes according to 16 different metadata such as disease, ecosystem, habitat, oxygen requirement, temperature, metabolism, etc.
It offers a very fast program CGC-Finder to identify CAZyme gene clusters (CGCs). The identified CGCs are presented in a CGC page.
FUTURE WORK
In the next release of dbCAN-seq, we will also include 700+ fungal genomes and 5,000+ metagenomes. We plan to update the database once a year to include newly defined CAZyme families, newly sequenced genomes and add new functions/features.
It is our hope that dbCAN-seq will become a primary website, where users from various research fields (genomics, carbohydrate, bioenergy, plant disease, food security, human microbiome and ecology) can quickly and easily download and browse the most comprehensive microbial CAZyme sequence and annotation data. Its function will be complementary to our dbCAN web server, which focuses on providing CAZyme online prediction service, and the CAZy database, which focuses on CAZyme family classification and nomenclature.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the Department of Computer Science of NIU for providing free access to the Linux computing cluster Gaea and Hartley. We also thank our lab members for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (NSF) CAREER award [DBI-1652164], National Institutes of Health (NIH) AREA award [1R15GM114706], Research & Artistry Award of the NIU [2016-YIN] to Y.Y.; National Natural Science Foundation of China [31728013 to Y.Y. and H.Z.] (in part). Funding for open access charge: NSF CAREER award [DBI-1652164].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B.. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014; 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coutinho P.M., Stam M., Blanc E., Henrissat B.. Why are there so many carbohydrate-active enzyme-related genes in plants?. Trends Plant Sci. 2003; 8:563–565. [DOI] [PubMed] [Google Scholar]
- 3. El Kaoutari A., Armougom F., Gordon J.I., Raoult D., Henrissat B.. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013; 11:497–504. [DOI] [PubMed] [Google Scholar]
- 4. Zhao Z., Liu H., Wang C., Xu J.R.. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013; 14:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rogowski A., Briggs J.A., Mortimer J.C., Tryfona T., Terrapon N., Lowe E.C., Basle A., Morland C., Day A.M., Zheng H. et al. . Glycan complexity dictates microbial resource allocation in the large intestine. Nat. Commun. 2015; 6:7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cockburn D.W., Koropatkin N.M.. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J. Mol. Biol. 2016; 428:3230–3252. [DOI] [PubMed] [Google Scholar]
- 7. Yin Y., Mao X., Yang J., Chen X., Mao F., Xu Y.. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012; 40:W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park B.H., Karpinets T.V., Syed M.H., Leuze M.R., Uberbacher E.C.. CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology. 2010; 20:1574–1584. [DOI] [PubMed] [Google Scholar]
- 9. Busk P.K., Pilgaard B., Lezyk M.J., Meyer A.S., Lange L.. Homology to peptide pattern for annotation of carbohydrate-active enzymes and prediction of function. BMC Bioinformatics. 2017; 18:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ekstrom A., Taujale R., McGinn N., Yin Y.. PlantCAZyme: a database for plant carbohydrate-active enzymes. Database. 2014; 2014:bau079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conners S.B., Mongodin E.F., Johnson M.R., Montero C.I., Nelson K.E., Kelly R.M.. Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species. FEMS Microbiol. Rev. 2006; 30:872–905. [DOI] [PubMed] [Google Scholar]
- 12. Freitas S., Hatosy S., Fuhrman J.A., Huse S.M., Welch D.B., Sogin M.L., Martiny A.C.. Global distribution and diversity of marine Verrucomicrobia. ISME J. 2012; 6:1499–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ward N.L., Challacombe J.F., Janssen P.H., Henrissat B., Coutinho P.M., Wu M., Xie G., Haft D.H., Sait M., Badger J. et al. . Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 2009; 75:2046–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flint H.J., Scott K.P., Duncan S.H., Louis P., Forano E.. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012; 3:289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chae L., Kim T., Nilo-Poyanco R., Rhee S.Y.. Genomic signatures of specialized metabolism in plants. Science. 2014; 344:510–513. [DOI] [PubMed] [Google Scholar]
- 16. Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R. et al. . CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017; 45:D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petersen T.N., Brunak S., von Heijne G., Nielsen H.. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011; 8:785–786. [DOI] [PubMed] [Google Scholar]
- 20. Juncker A.S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., Krogh A.. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. Soc. 2003; 12:1652–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001; 305:567–580. [DOI] [PubMed] [Google Scholar]
- 22. Yan R., Xu D., Yang J., Walker S., Zhang Y.. A comparative assessment and analysis of 20 representative sequence alignment methods for protein structure prediction. Scientific Rep. 2013; 3:2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Larsbrink J., Rogers T.E., Hemsworth G.R., McKee L.S., Tauzin A.S., Spadiut O., Klinter S., Pudlo N.A., Urs K., Koropatkin N.M. et al. . A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014; 506:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sonnenburg E.D., Zheng H., Joglekar P., Higginbottom S.K., Firbank S.J., Bolam D.N., Sonnenburg J.L.. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010; 141:1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ravcheev D.A., Godzik A., Osterman A.L., Rodionov D.A.. Polysaccharides utilization in human gut bacterium Bacteroides thetaiotaomicron: comparative genomics reconstruction of metabolic and regulatory networks. BMC Genomics. 2013; 14:873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Terrapon N., Henrissat B.. How do gut microbes break down dietary fiber?. Trends Biochem. Sci. 2014; 39:156–158. [DOI] [PubMed] [Google Scholar]
- 27. Terrapon N., Lombard V., Gilbert H.J., Henrissat B.. Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics. 2015; 31:647–655. [DOI] [PubMed] [Google Scholar]
- 28. Saier M.H. Jr, Reddy V.S., Tsu B.V., Ahmed M.S., Li C., Moreno-Hagelsieb G.. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 2016; 44:D372–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kilic S., White E.R., Sagitova D.M., Cornish J.P., Erill I.. CollecTF: a database of experimentally validated transcription factor-binding sites in Bacteria. Nucleic Acids Res. 2014; 42:D156–D160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sierro N., Makita Y., de Hoon M., Nakai K.. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008; 36:D93–D96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muniz-Rascado L., Garcia-Sotelo J.S., Alquicira-Hernandez K., Martinez-Flores I., Pannier L., Castro-Mondragon J.A. et al. . RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016; 44:D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 33. Chen I.A., Markowitz V.M., Chu K., Palaniappan K., Szeto E., Pillay M., Ratner A., Huang J., Andersen E., Huntemann M. et al. . IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017; 45:D507–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.