Abstract
Large-scale sequencing studies discovered substantial genetic variants occurring in enhancers which regulate genes via long range chromatin interactions. Importantly, such variants could affect enhancer regulation by changing transcription factor bindings or enhancer hijacking, and in turn, make an essential contribution to disease progression. To facilitate better usage of published data and exploring enhancer deregulation in various human diseases, we created DiseaseEnhancer (http://biocc.hrbmu.edu.cn/DiseaseEnhancer/), a manually curated database for disease-associated enhancers. As of July 2017, DiseaseEnhancer includes 847 disease-associated enhancers in 143 human diseases. Database features include basic enhancer information (i.e. genomic location and target genes); disease types; associated variants on the enhancer and their mediated phenotypes (i.e. gain/loss of enhancer and the alterations of transcription factor bindings). We also include a feature on our website to export any query results into a file and download the full database. DiseaseEnhancer provides a promising avenue for researchers to facilitate the understanding of enhancer deregulation in disease pathogenesis, and identify new biomarkers for disease diagnosis and therapy.
INTRODUCTION
More than 80% of genetic variants associated with complex traits were observed in noncoding regions (1). Specially, disease-associated variants are significantly enriched in enhancer regulatory elements which regulate target genes via long range chromatin interactions (2). Since the first disease-associated enhancer was identified in Hirschsprung disease as early as 2005 (3), large-scale sequencing studies, to date, have discovered substantial somatic mutations occurring in enhancers (4). Importantly, emerging evidence suggested that genetic alterations of enhancers could make an essential contribution to disease progression (5–8). Single nucleotide variants (SNVs) and small insertions or deletions (indels) could lead to a gain of a de novo enhancer or loss of an existed enhancer, and change the transcription factor bindings in the genome. For example, Marc et al. found a de novo super-enhancer formed through a somatic mutation, which created a binding site for MYB and in turn activated the oncogene TAL1 expression in T-cell acute lymphoblastic leukemia (9). Moreover, structure-related variants, such as copy number variants (CNVs) and rearrangements, could change the output of enhancer or affect the enhancer regulation by enhancer hijacking, altering the transcriptional regulation of downstream key target genes. For example, Lori et al. found that an aberrant enhancer was hijacked by MYB-QKI rearrangement, which driven MYB overexpression and tumorigenicity in glioma (10).
At present, hundreds of thousands of enhancers have been identified by ENCODE (11), Roadmap (12), FANTOM5 (13) and a very recent project, Blueprint/IHEC (14). Undoubtedly, exploring disease-associated enhancers will open a brand new view of pathophysiology. Indeed, the publications deciphering the roles of enhancers in disease pathogenesis increased in an exponential manner, especially in recent years. However, none of the developed enhancer-related databases considered the enhancer-disease associations, such as VISTA Enhancer Browser (15), EnhancerAtlas (16), dbSUPER (17) and SEA (18). Consequently, the time has arrived to gather the current knowledge of disease-associated enhancers, which are still hidden in a large amount of dispersed literatures, allowing researchers to expand their view to disease pathogenesis at the enhancer levels.
We therefore developed DiseaseEnhancer, a novel database that provides for the first time a manually curated collection of disease-associated enhancers. The current release of DiseaseEnhancer includes 847 disease-associated enhancers in 143 human diseases. We expect to see an increase of instances over the years as the rapidly increasing interest in enhancers-disease associations. DiseaseEnhancer makes all disease-associated enhancer information publicly available in one location, providing an important and live-updated resource that facilitates the understanding of regulatory mechanisms in disease pathogenesis.
DATA COLLECTION, PROCESSING AND IMPLEMENTATION
Using a list of keywords, such as ‘enhancer’, ‘regulatory element’, ‘risk loci’, ‘mutation’, ‘copy number’, ‘variant’, ‘predisposition’, ‘susceptibility’, ‘cancer’, ‘tumor’ and ‘disease’, we obtained 1866 publications from PubMed. We manually examined the abstracts to obtain the pertinent publications that describe enhancer-disease associations. By scrutinizing the full texts of these publications, we extracted the information of disease-associated enhancers that were verified strictly by a series of experiments, including validation of enhancers, phenotypic/mechanistic characterization of relevant genetic alterations, and determination of targets of enhancers. Specifically, the enhancers must be verified by active chromatin marks (e.g. H3K4me1 or H3K27ac); the relevant genetic variants could contribute to disease phenotype or affect the binding of important transcription factors; the target genes should be identified based on the enhancer-promoter interactions by either experimental (e.g. capturing chromosome conformation (3C) (19), chromosome conformation capture-on-chip (4C) (20) and high-throughput chromosome conformation capture (Hi-C) (21)) or computational methods (e.g. integrated method for predicting enhancer targets (IM-PET) (22)). For example, Swneke et al. verified an enhancer associated with breast cancer (23). They first identified enhancers using DNase-seq data in breast cancer cell lines, and then determined the target genes of the enhancers using Cross-Cell type Correlation in DNaseI hypersensitivity (C3D) method (24), which were further validated by chromatin interaction analysis with paired-end-tag sequencing (ChIA-PET) (25) and 3C-qPCR (19). Using somatic mutation data from the cancer genome atlas (TCGA) (26), Swneke et al. found a mutation (chr6:151955219, T>G) occurring in an enhancer of ESR1 (chr6:151953109–151955347) and verified the association of the mutation with breast cancer risk by cell viability assays. Furthermore, they validated that the mutation can result in a loss of GATA3 binding and increasing of enhancer activity using chromatin immunoprecipitation (ChIP) (27) and luciferase reporter assays (28).
During the extraction of genomic locations of disease-associated enhancers from the original publications, we found that many publications did not provide the detailed information about the genomic locations. Only the locations of relevant genetic variations were available. In such cases, we mapped the relevant genetic variations to known annotated enhancers, which were derived from the union of enhancers from Roadmap (12) and FANTOM5 (13) databases, using the GenomicRanges package in Bioconductor (29). Of note, some large genetic alterations harboring multiple known enhancers were excluded. When the genetic variations (SNVs and indels) were not mapped to any known enhancers, the ±1 kb regions around the genetic variations were used as the disease-associated enhancers. All the genomic locations were unified in hg19 coordinates to facilitate further applications.
Besides, other detailed information including enhancer-associated variant type, location, mode and consequence and the enhancer target gene were also gathered. Variant consequence generally includes the following information: whether the relevant variant can change the enhancer activity (i.e. gain/loss of enhancer, increasing/decreasing of enhancer activity); which TFs are affected by the variant when binding to the disease-associated enhancer (i.e. gain/loss of a TF motif or increasing/decreasing of a TF binding).
In some cases, the information of disease-associated enhancers recorded in original publications was difficult to be gathered, as they were presented in distinct and unexportable formats, such as tables encoded into PDF documents and figures in the main text as well as supplementary materials. To collect the information, we carefully scrutinized the corresponding figures and tables in the full text and the supplementary data, and contacted with authors as necessary. In addition, we merged the entries with the same genomic location and variants to avoid duplications within the database. All the curation were done and double-checked by multiple researchers.
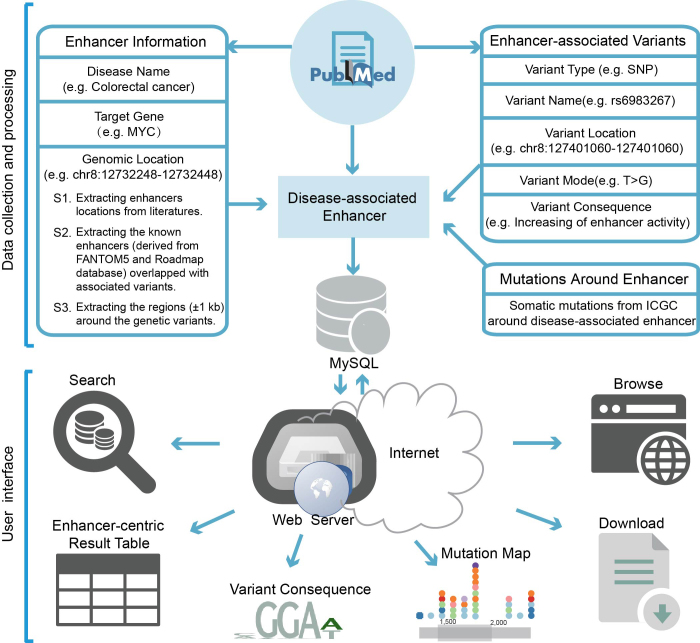
We also assess the mutation frequency of disease-associated enhancers using mutation profiles from International Cancer Genome Consortium (ICGC) (30). Genetic mutations within +/-5 kb regions around the disease-associated enhancers were identified using bedtools (31) to further consolidate the understanding of disease-associated enhancers. Finally, the disease-associated enhancers and all the related information were loaded into MySQL. Figure 1 schematically illustrates the general workflow and features of the database.
Figure 1.
Overview of disease-associated enhancer collection, annotation and DiseaseEnhancer database features.
DiseaseEnhancer is freely available to the research community at http://biocc.hrbmu.edu.cn/DiseaseEnhancer/ and requires no registration or login. We have tested it in Mozilla Firefox, Google Chrome and Apple Safari browsers. We will update the database and website in regular intervals per year depending on the number of new researches focusing on discovering novel disease-associated enhancers.
DATABASE STATISTICS
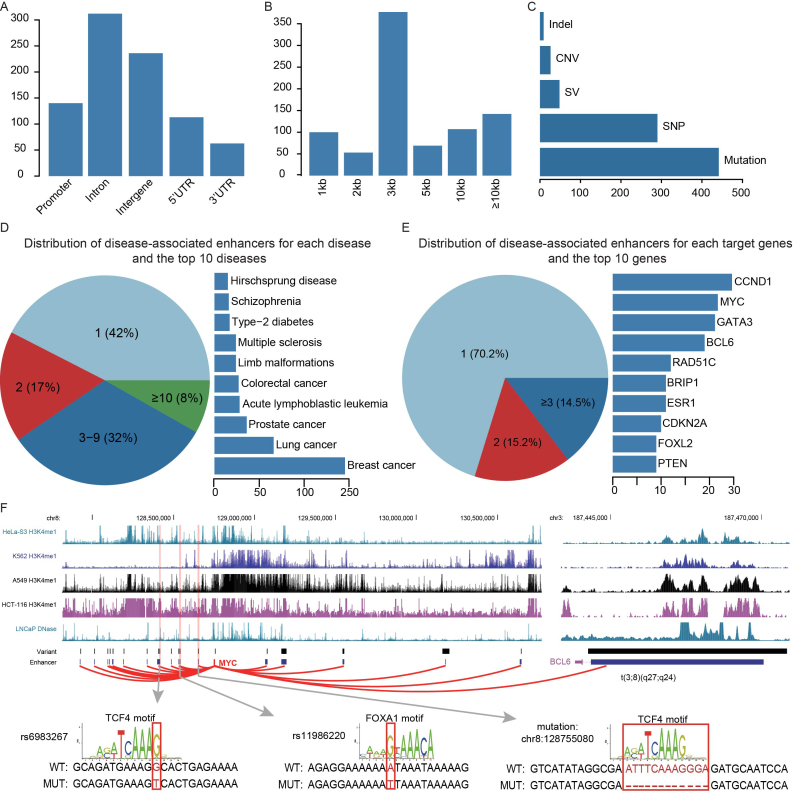
As of July 2017, DiseaseEnhancer consists of 847 disease-associated enhancers in 143 human diseases. The disease-associated enhancers were mainly located in intron (36.8%), intergenic (27.9%) and 3′UTR (5.4%), and nearly 18% enhancers overlapped with promoters and 5′UTR (Figure 2A). Of note, 44.8% of disease-associated enhancers overlapping promoters are super enhancers, which are large clusters of enhancers driving expression of genes responsible for cell identity and thus span long genomic regions. Another possible explanation for the overlap between disease-associated enhancers and promoters is due to the enhancer-like action of promoters in higher chromosomal organization (25,32). Most of the disease-associated enhancers were smaller than 3kb (62.6%), on the median of 2kb (Figure 2B). In addition, 91.2% of the curated disease-associated enhancers were affected by mutations, indels and SNPs. Conversely, only a small number of disease-associated enhancers within CNVs and structure variants(SVs, including inversion, translocation and duplication) were identified, which probably reflects the difficulty of identifying the causal disease-associated enhancers in the jumble of passenger enhancers affected by a large alteration (Figure 2C). The top 10 diseases ranked by the number of associated enhancers include breast cancer (n = 245), lung cancer (n = 66), prostate cancer (n = 36), acute lymphoblastic leukemia (n = 28), colorectal cancer (n = 27), limb malformation (n = 24), multiple sclerosis (n = 24), type-2 diabetes (n = 17), schizophrenia (n = 16) and hirschsprung disease (n = 15), which are represented by 58.8% of the curated disease-associated enhancers (Figure 2D). The majority (132 out of 143 or 92.3%) of diseases in DiseaseEnhancer are found to be associated with less than 10 enhancers, indicating the further exploring of disease-associated enhancers is essential for understanding disease progression. A total of 308 target genes were regulated by the disease-associated enhancers, 29.8% (92 out of 308) of these genes were regulated by more than a single enhancer in the diseases (Figure 2E). One of the top target gene ranked by the number of associated enhancers, MYC, was affected by 22 disease-associated enhancers in multiple diseases by various mechanisms (Figure 2F). For example, MYC was aberrantly activated by a de novo enhancer created by a SNP (rs6983267) in colorectal cancer (33), while in B-cell lymphoma, by a super enhancer hijacked by a rearrangement (t(3;8)(q27;q24)) (34).
Figure 2.
Statistics of DiseaseEnhancer. (A) The distribution of disease-associated enhancers in genomic elements. (B) The width of disease-associated enhancers. (C) The distribution of genetic variation types. (D) Majority of the diseases harbor less than 10 disease-associated enhancers (left). And the top 10 diseases ranked by associated enhancers are shown (right). (E) Part of the target genes were regulated by more than a single enhancer (left). And the top 10 genes ranked by associated enhancers are shown (right). (F) The twenty two disease-associated enhancers affected MYC in eight diseases. The sequence of three enhancers and the affected transcription factor motifs are also shown.
WEB INTERFACE
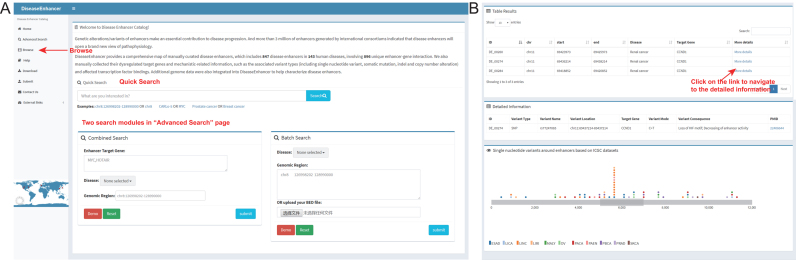
DiseaseEnhancer's public user interface allows intuitive browsing and searching of all enhancer-disease associations in the database (Figure 3A). The home page of the website provides a quick search utility which can be used to query the database for genomic region, genes of interest as well as disease names. The ‘Advanced Search’ provides multiple options to users for the customized search of interested region-gene-disease associations. Notably, the target gene searches will also return all entries that contain the characters specified. For instance, searching for ‘MYC’ will also return the entries containing ‘CARLo-5, MYC’. In addition, a batch search of disease-associated enhancers is also provided in this page. Users can obtain the matched disease-associated enhancers in the regions of interest by inputting certain genomic regions or uploading a file in BED3 format.
Figure 3.
An illustration of DiseaseEnhancer. (A) Browser shots of the database. A user can simply query the database by typing a gene symbol, a genomic location or a disease name of interest into the ‘Quick Search’ box in the home page. In the ‘Advanced Search’ page, users can search for disease-associated enhancers of interest by combining gene symbol, genomic location and disease name options in the ‘Combined Search’ module. A ‘Batch Search’ module is also built in this page for search for multiple enhancers of interest in a specific disease at one time. Users can browse the disease-associated enhancers in a specific disease by selecting disease name in the ‘Browse’ page. (B) Results of a search for CCND1 in renal cancer. Disease-associated enhancers of CCND1 in renal cancer are presented in a brief result table. After clicking the ‘More details’ link for a specific enhancer, the detailed annotations on the enhancer will be shown, including the target genes, the associated genetic variants, changing modes and variant consequences, and the somatic mutation map of around the enhancer.
The query result is presented as a responsive table, which contains the basic information of disease-associated enhancers, including ID maintained by our database, genomic location, associated disease as well as target gene (Figure 3B). After clicking the ‘More details’ link for a specific disease-associated enhancer, user could obtain the details about the variations on the enhancer region including the enhancer-associated variant type, name, location, mode (such as C>T), consequence (such as change of TF binding and the enhancer activity), target gene and links to the reference. In addition, the responsive table provides several useful features. Firstly, users can further filter the result in the table by typing terms in the ‘Search’ box at the top-right of the table; and only the entries matching the term will be displayed. Secondly, each column could be ranked in ascending or descending by clicking the arrows in the column header. Third, the disease-associated enhancers could be exported to a tab-separated file by clicking the ‘Excel’ button beside the ‘Search’ box.
Moreover, the database can be browsed for each disease by clicking the ‘Browse’ tab on the left-side navigation menu. In this page, users could quickly obtain all the curated enhancers associated with a disease of interest by selecting the disease name in a pull-down menu. In addition, DiseaseEnhancer also provides a detailed tutorial for the usage of the database in the ‘Help’ page. The download version of the full dataset is available as a tab-delimited file on the ‘Download’ page of the website and submission of new or updated information based peer-reviewed publications is available on the ‘Submit’ page.
SUMMARY AND PROSPECTICTIVE WORKS
New trends are starting to emerge in deciphering disease initiation, progression and therapeutics with considering both genome and epigenome. Enhancers have become a type of key elements to decipher the dysregulated mechanism, linking aberrant genome and epigenome, underlying diseases. Recent researches, such as the evolution trajectory of acute lymphoblastic leukemia (35) and epigenetic therapy in cancers (36), underscored that enhancers will be a new star in cancer researches, making a great blueprint for disease researches in the future. Aggregating the information of disease-associated enhancers, thus, is an essential step. However, there is no such database available now, although several enhancer-related databases have been developed, such as VISTA Enhancer Browser (15) and EnhancerAtlas (16), focusing on the catalog of various enhancers. Here, we developed DiseaseEnhancer, aiming at integrating information on the enhancers underpinning diseases, and in such endeavor, closing the disease-enhancer gap.
DiseaseEnhancer is a first repository that provides a comprehensive collection of experimentally validated disease-associated enhancers for the scientific community. It currently contains 847 disease-associated enhancers in 143 human diseases. Considering genetic variants are the basis of diseases, DiseaseEnhancer also provides a mutation map plot to show the mutations mapped in the disease-associated enhancers to help understand the roles of enhancers in diseases. For example, by searching DiseaseEnhancer using both ‘CCND1’ and ‘Renal cancer’, we found that recurrent mutations occurred in the enhancer of CCND1 (DE_00274) in cancers, implying the recurrent mutations may also affect the enhancer function.
Utilizing the data in DiseaseEnhancer, a network of diseases and enhancers were constructed (Figure 4). 76.1% of the disease-associated enhancers were rare and restricted to one disease type, probably due to the high specificity of enhancers. Meanwhile, 23.9% of the disease-associated enhancers were frequently changed in diverse types of disease. Among these enhancers, seven were shared by multiple sclerosis, lupus, ulcerative colitis, Celiac disease, Rheumatoid arthritis and Crohn disease. All of these disorders exhibited an autoimmune phenotype. The target genes of these disease-associated enhancers, including PFKFB3, ADO, CD40, ICOSLG, IRF5 and FAM167A, were all involved in immune functions, and enriched in “defense response to virus" (P-value<0.05, hypergeometric test). Furthermore, several enhancers were shared in similar diseases. For example, three enhancers are consistently linked with bipolar disorder and schizophrenia. Interestingly, in cancers, several common enhancers were also observed. Abnormalities in these enhancers could affected some well-known cancer genes, such as MYC, CDKN2A, SOX9 and IL2RA, implying a similar underlying mechanism.
Figure 4.
Network of enhancers and diseases. Only the diseases which harbor more than 5 associated enhancers and their neighbors are shown. Red circle represents disease, blue circle represents disease-associated enhancer, green circle represents target gene.
The important roles of enhancers in disease are attracting more and more attention. With the rapidly researching on disease in an epigenetic view, more experimentally supported enhancer-disease associations are expected to be published in the future and these data will be integrated into the DiseaseEnhancer. Furthermore, with the increasing of new genomic and epigenomic data both derived from patient, we hope that we can construct a reliable supervised predictive model using the data in DiseaseEnhancer as positive set for identifying the potential disease-enhancer associations, and integrate the model into the DiseaseEnhancer database in the future. In addition, we also plan to add an integrative visualization module that integrates gene and enhancer annotation, genetic variations, transcription factor binding sites, comprehensive chromatin interactions and gene expression to help mining the underlying mechanism in disease progression.
In summary, DiseaseEnhancer is a comprehensive collection of experimentally validated disease-associated enhancers. It will be regularly growing with the new researches, new data sources, and the interest of a growing community of users. The careful curation and the attention paid to keeping track of the related information, make of DiseaseEnhancer a platform of choice to support further research in disease.
FUNDING
National High Technology Research and Development Program of China (863 Program) [2014AA021102]; National Program on Key Basic Research Project (973 Program) [2014CB910504]; National Natural Science Foundation of China [91439117, 61473106, 61573122]; China Postdoctoral Science Foundation [2016M600260]; Wu lien-teh youth science fund project of Harbin Medical University [WLD-QN1407]; Special funds for the construction of higher education in Heilongjiang Province [UNPYSCT-2016049]; Heilongjiang Postdoctoral Foundation [LBH-Z16098]. Funding for open access charge: National High Technology Research and Development Program of China (863 Program) [2014AA021102]; National Program on Key Basic Research Project (973 Program) [2014CB910504]; National Natural Science Foundation of China [91439117, 61473106, 61573122]; China Postdoctoral Science Foundation [2016M600260]; Wu lien-teh youth science fund project of Harbin Medical University [WLD-QN1407]; Special funds for the construction of higher education in Heilongjiang Province [UNPYSCT-2016049]; Heilongjiang Postdoctoral Foundation [LBH-Z16098].
Conflict of interest statement. None declared.
REFERENCES
- 1. Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012; 337:1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Farh K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015; 518:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grice E.A., Rochelle E.S., Green E.D., Chakravarti A., McCallion A.S.. Evaluation of the RET regulatory landscape reveals the biological relevance of a HSCR-implicated enhancer. Hum. Mol. Genet. 2005; 14:3837–3845. [DOI] [PubMed] [Google Scholar]
- 4. Melton C., Reuter J.A., Spacek D.V., Snyder M.. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat. Genet. 2015; 47:710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu N.Q., Ter Huurne M., Nguyen L.N., Peng T., Wang S.Y., Studd J.B., Joshi O., Ongen H., Bramsen J.B., Yan J. et al. The non-coding variant rs1800734 enhances DCLK3 expression through long-range interaction and promotes colorectal cancer progression. Nat. Commun. 2017; 8:14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pasquali L., Gaulton K.J., Rodriguez-Segui S.A., Mularoni L., Miguel-Escalada I., Akerman I., Tena J.J., Moran I., Gomez-Marin C., van de Bunt M. et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 2014; 46:136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jager R., Migliorini G., Henrion M., Kandaswamy R., Speedy H.E., Heindl A., Whiffin N., Carnicer M.J., Broome L., Dryden N. et al. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat. Commun. 2015; 6:6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doan R.N., Bae B.I., Cubelos B., Chang C., Hossain A.A., Al-Saad S., Mukaddes N.M., Oner O., Al-Saffar M., Balkhy S. et al. Mutations in human accelerated regions disrupt cognition and social behavior. Cell. 2016; 167:341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mansour M.R., Abraham B.J., Anders L., Berezovskaya A., Gutierrez A., Durbin A.D., Etchin J., Lawton L., Sallan S.E., Silverman L.B. et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014; 346:1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bandopadhayay P., Ramkissoon L.A., Jain P., Bergthold G., Wala J., Zeid R., Schumacher S.E., Urbanski L., O’Rourke R., Gibson W.J. et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat. Genet. 2016; 48:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roadmap Epigenomics C., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lizio M., Harshbarger J., Shimoji H., Severin J., Kasukawa T., Sahin S., Abugessaisa I., Fukuda S., Hori F., Ishikawa-Kato S. et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015; 16:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fernandez J.M., de la Torre V., Richardson D., Royo R., Puiggros M., Moncunill V., Fragkogianni S., Clarke L., Consortium B., Flicek P. et al. The BLUEPRINT data analysis portal. Cell Syst. 2016; 3:491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Visel A., Minovitsky S., Dubchak I., Pennacchio L.A.. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007; 35:D88–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao T., He B., Liu S., Zhu H., Tan K., Qian J.. EnhancerAtlas: a resource for enhancer annotation and analysis in 105 human cell/tissue types. Bioinformatics. 2016; 32:3543–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khan A., Zhang X.. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016; 44:D164–D171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei Y., Zhang S., Shang S., Zhang B., Li S., Wang X., Wang F., Su J., Wu Q., Liu H. et al. SEA: a super-enhancer archive. Nucleic Acids Res. 2016; 44:D172–D179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dekker J., Rippe K., Dekker M., Kleckner N.. Capturing chromosome conformation. Science. 2002; 295:1306–1311. [DOI] [PubMed] [Google Scholar]
- 20. Splinter E., de Wit E., Nora E.P., Klous P., van de Werken H.J., Zhu Y., Kaaij L.J., van Ijcken W., Gribnau J., Heard E. et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011; 25:1371–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Belton J.M., McCord R.P., Gibcus J.H., Naumova N., Zhan Y., Dekker J.. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012; 58:268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He B., Chen C., Teng L., Tan K.. Global view of enhancer-promoter interactome in human cells. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2191–E2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bailey S.D., Desai K., Kron K.J., Mazrooei P., Sinnott-Armstrong N.A., Treloar A.E., Dowar M., Thu K.L., Cescon D.W., Silvester J. et al. Noncoding somatic and inherited single-nucleotide variants converge to promote ESR1 expression in breast cancer. Nat. Genet. 2016; 48:1260–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. et al. The accessible chromatin landscape of the human genome. Nature. 2012; 489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012; 148:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cancer Genome Atlas Research, N. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M.. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Collas P. The current state of chromatin immunoprecipitation. Mol. Biotechnol. 2010; 45:87–100. [DOI] [PubMed] [Google Scholar]
- 28. Gould S.J., Subramani S.. Firefly luciferase as a tool in molecular and cell biology. Anal. Biochem. 1988; 175:5–13. [DOI] [PubMed] [Google Scholar]
- 29. Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang J., Baran J., Cros A., Guberman J.M., Haider S., Hsu J., Liang Y., Rivkin E., Wang J., Whitty B. et al. International Cancer Genome Consortium Data Portal—a one-stop shop for cancer genomics data. Database. 2011; 2011:bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park S.M., Choi E.Y., Bae M., Choi J.K., Kim Y.J.. A long-range interactive DNA methylation marker panel for the promoters of HOXA9 and HOXA10 predicts survival in breast cancer patients. Clin. Epigenet. 2017; 9:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tuupanen S., Yan J., Turunen M., Gylfe A.E., Kaasinen E., Li L., Eng C., Culver D.A., Kalady M.F., Pennison M.J. et al. Characterization of the colorectal cancer-associated enhancer MYC-335 at 8q24: the role of rs67491583. Cancer Genet. 2012; 205:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryan R.J., Drier Y., Whitton H., Cotton M.J., Kaur J., Issner R., Gillespie S., Epstein C.B., Nardi V., Sohani A.R. et al. Detection of enhancer-associated rearrangements reveals mechanisms of oncogene dysregulation in B-cell lymphoma. Cancer Discov. 2015; 5:1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Corces M.R., Buenrostro J.D., Wu B., Greenside P.G., Chan S.M., Koenig J.L., Snyder M.P., Pritchard J.K., Kundaje A., Greenleaf W.J. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 2016; 48:1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dawson M.A. The cancer epigenome: concepts, challenges, and therapeutic opportunities. Science. 2017; 355:1147–1152. [DOI] [PubMed] [Google Scholar]