Abstract
DifferentialNet is a novel database that provides users with differential interactome analysis of human tissues (http://netbio.bgu.ac.il/diffnet/). Users query DifferentialNet by protein, and retrieve its differential protein–protein interactions (PPIs) per tissue via an interactive graphical interface. To compute differential PPIs, we integrated available data of experimentally detected PPIs with RNA-sequencing profiles of tens of human tissues gathered by the Genotype-Tissue Expression consortium (GTEx) and by the Human Protein Atlas (HPA). We associated each PPI with a score that reflects whether its corresponding genes were expressed similarly across tissues, or were up- or down-regulated in the selected tissue. By this, users can identify tissue-specific interactions, filter out PPIs that are relatively stable across tissues, and highlight PPIs that show relative changes across tissues. The differential PPIs can be used to identify tissue-specific processes and to decipher tissue-specific phenotypes. Moreover, they unravel processes that are tissue-wide yet tailored to the specific demands of each tissue.
INTRODUCTION
Proteins, the main building blocks of living cells, carry out their functions by interacting with other molecules and particularly with other proteins. Analysis of their interactions therefore unravels their molecular functions and roles in health and disease (1–3). Realizing the importance of their interactions, many studies conducted large-scale screens for interaction detection by using various experimental techniques (4–7). In human, which is the focus of DifferentialNet, over 240 000 protein–protein interactions (PPIs) between >20 000 human proteins have been discovered to date (8). Combining all known PPIs into one network, denoted interactome, results in a huge, dense, ‘hair-ball’ network.
Large-scale PPI detection screens tend to provide limited physiological context. Yeast-two-hybrid screens, for example, detect PPIs within yeast cells, and affinity-based assays are typically carried in a single condition and are not repeated across conditions (3,9). Yet in biological settings PPIs are context-sensitive. The human body is composed of many tissues and cell types that differ from each other by the levels of genes and proteins that they express (10–13). Human proteins, therefore, frequently interact with different partners in distinct tissues and cell types (14,15). For example, the protein DAG1 is expressed throughout the human body, yet interacts with the protein CAV3 only in the subset of tissues that express CAV3, such as muscle and heart, and not in other tissues.
In recent years, several studies incorporated tissue and cell type contexts into the human interactome. The general approach for creating a tissue interactome is to penalize PPIs between pair-mates that are lowly-expressed or undetectable in that tissue, or remove them completely from the network (14,16–19). Such tissue interactomes were used in several applications and outperformed the global interactome in illuminating the molecular basis of hereditary diseases (20) or in prioritizing candidate disease genes (16–18,21,22).
To get a better understanding of the processes that separate a specific tissue from other tissues, we should consider the quantitative changes in gene expression and how they relate to PPIs. Rather than ask ‘which interactions are most expressed in a certain tissue?’ we should ask ‘which interactions are most altered between tissues?’ (23). This differential network approach aims to identify PPIs that are differentially relevant in the tissue or condition of interest. The power of this approach was nicely illustrated by the usage of differential epistasis mapping for understanding DNA-damage response in yeast (24). The genetic interaction network that was measured in yeast cells that were perturbed with a DNA-damaging agent did not show a clear DNA-damage signature; only upon focusing on interactions that were altered significantly between this network and the network measured in unperturbed cells, the DNA-damage signature emerged.
Several tools offer schemes for differential network analysis. The Differential Network Analysis (DNA) package analyzes differential co-expression based on microarray data. It calculates expression correlations between genes, and highlights gene pairs with altered co-regulation between conditions (25). The Diffany tool uses an interaction ontology to infer differential networks and functional roles (26). The DINA framework identifies gene pairs whose co-expression is condition-specific based on microarray data, and predicts transcriptional regulators that may be responsible for their co-regulation (27). DyNet is a Cytoscape application that computes the variance of nodes and interactions in different networks, and highlights those that are highly variable (28). DINGO is a framework that estimates topological differences between networks and highlights sub-networks that are altered (29). This was used to elucidate cancer development pathways. These various tools are available as desktop applications, and require that users gather expression-data and/or PPI data on their own. Furthermore, some of the tools fit microarray data, which are becoming less common.
Here, we present DifferentialNet, a novel web-based database for differential network analysis of human tissue interactomes. To this end, we integrated current data of experimentally-detected PPIs with data of gene expression across tissues, to obtain a differential view of the human interactome. Data of gene expression across tissues was gathered by two major initiatives, The Genotype-Tissue Expression (GTEx) consortium (30) and the Human Protein Atlas (HPA) (11), that applied RNA-sequencing to samples from 42 and 29 tissues, respectively. DifferentialNet associates each PPI with a tissue-specific score that reflects how its likelihood changed in the selected tissue relative to other tissues, based on the expression levels of the pair-mates across tissues. Users query DifferentialNet by protein and tissue, and obtain a graphical network that highlights the differential interactions of the protein in that tissue along with additional information regarding the presented interactions. Users can interactively alter the highlighted interactions and toggle between different tissues for comparative analyses. Thus, DifferentialNet greatly facilitates analyses of the altered roles of human proteins and their interactions across tissues.
RESULTS
The DifferentialNet approach
DifferentialNet synergizes between large-scale data of PPI and expression profiles across tissues to create a novel database that points to PPIs that are up- or down-regulated in a selected human tissue relative to all other human tissues. To compute differential PPIs, we gathered PPIs from four major PPI databases, BioGrid (8), DIP (31), MINT (32) and IntAct (33), and consolidated them by using the MyProteinNet web-server (34). This resulted in a global interactome that contained 200,183 PPIs between 16,532 human proteins. The use of MyProteinNet guaranteed that only PPIs detected by established experimental methods were included. Expression data were gathered from GTEx, which included 421 samples from 42 tissues, and from HPA, which included 192 samples from 29 tissues (see Methods). Only genes that were expressed above a certain threshold in at least one tissue were included (see Methods). This resulted in interactomes that contained 134,223 PPIs between 13,523 human proteins for GTEx, and interactomes that contained 137,613 PPIs between 13,613 human proteins for HPA.
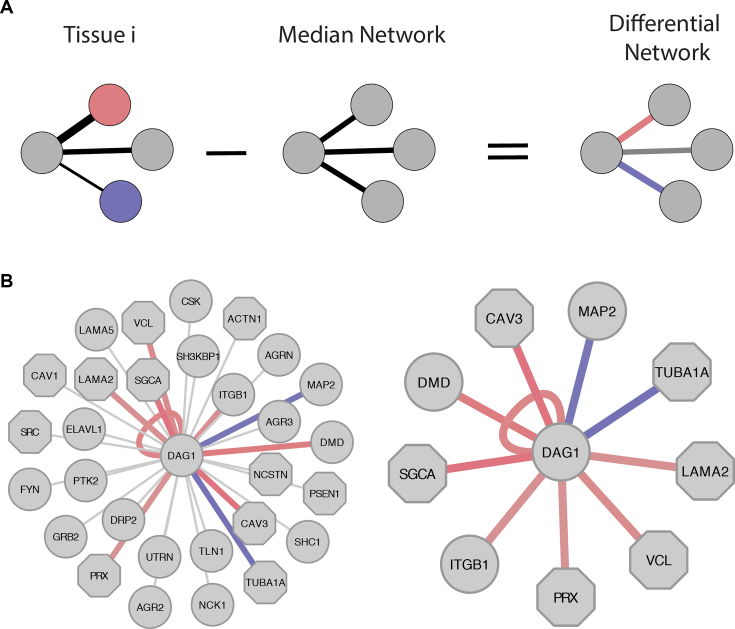
For each tissue in each dataset, we created its differential interactome. This was achieved as follows. First, we assigned each interaction with a tissue-specific score that reflects the likelihood of the interaction, and is based on multiplying the expression levels of the interacting pair-mates (see Methods). This implies that interactions between highly expressed partners were scored high. We repeated this procedure across all tissues, and recorded for each interaction its median score across all tissues. The differential tissue-specific score of an interaction was then computed as the difference between its tissue-specific score and its median score (Figure 1A). By this scoring method, interactions between pair-mates that were similarly expressed across all tissues, i.e. unchanged, had low differential scores, even if the pair-mates were highly expressed. In contrast, interactions between pair-mates that had different expression levels across tissues were assigned positive or negative scores, according to whether they were up- or down-regulated in that tissue.
Figure 1.
DifferentialNet model and output. (A) A flowchart describing the scoring framework of DifferentialNet. The differential analysis starts with scoring tissue PPIs by the expression levels of the interacting genes. Then, a median interactome is created, such that the score of each PPI is set to its median score across all tissues. The median score of each interaction is then subtracted from its tissue score, to create a tissue-specific differential network. The differential scores represent whether the interaction is up-regulated (red edge) or down-regulated (blue edge) in the tissue. (B) The differential network view for the protein DAG1 in skeletal muscle based on GTEx. The network on the left shows DAG1 with all its PPIs, where up-regulated PPIs appear in red, down-regulated PPIs appear in blue, and unchanged PPIs are marked in gray. The network on the right shows only differential PPIs that score at 10th percentile (top 10th percentile in red and bottom 10th percentile in blue).
DifferentialNet usage
Users query DifferentialNet by a protein, a tissue and a resource (GTEx or HPA), and can search up to five proteins in one query. Proteins can be specified by using Ensembl gene ID, Entrez gene ID, or gene symbols. The full lists of proteins with PPIs in DifferentialNet are accessible from the DifferentialNet homepage. DifferentialNet also provides a ‘sample protein’ query that retrieves the differential PPIs of the human protein DAG1 (Figure 1B), and a ‘random protein’ query that retrieves the differential PPIs of a randomly selected protein and tissue.
DifferentialNet output includes a graphical network view that uses the Cytoscape.js plugin (35), and textual information of the output proteins and PPIs. The network denotes proteins as nodes and PPIs as edges, while highlighting their differential tissue scores: up-regulated interactions appear in red and down-regulated interactions appear in blue. DifferentialNet also offers an interactive user interface which allows users to set a percentile threshold for the differential scores. By this they can filter the displayed PPIs, so that only PPIs that score in the top percentile (e.g., top 10%) or in the bottom percentiles (e.g., bottom 10%) are shown. A sliding bar allows users to interactively set the percentile threshold to obtain an adjusted network. Another menu allows users to toggle between different tissues, or to display unfiltered interactions (Figure 1B). DifferentialNet also highlights disease-associated proteins by using a different node shape. Data of disease-association were gathered from the Online Mendelian Inheritance in Man (OMIM) (36) database.
The textual information is divided into tabs, each relating to a different type of data regarding the output. The ‘Properties’ tab lists the different gene identifiers and detection methods for selected proteins and PPIs. The ‘Gene Ontology’ (GO) tab provides the GO annotations (37) and description of selected proteins in the network, obtained from the MyGene.info (38) web-service. The ‘Differential’ tab lists the scores and percentiles of selected PPIs in every tissue. Lastly, the ‘Graph Options’ tab allows users to change the network layout, remove proteins from the network, expand the network from a selected protein, or display PPIs that are differential in few tissues or across many tissues. It also allows users to export the network in Cytoscape.JSON format for further analysis in Cytoscape.
SUMMARY
The DifferentialNet database provides differential interactomes for over 29 human tissues by scoring data of PPIs by data of gene expression according to user-defined parameters. The output of DifferentialNet highlights differential PPI sub-networks and disease genes. By this, DifferentialNet offers a novel means to illuminate protein functions, processes and phenotypes that are either preferred or disfavored in certain tissues. DifferentialNet functionality and user-friendly interface can accommodate new data of additional tissues and interactions as they become available. In face of the increasing density of the human interactome, tools like DifferentialNet that provide sparse and context-specific views into the interactome become ever more important in applied research of human phenotypes and diseases.
METHODS
Expression data sources
Tissue expression profiles were gathered from GTEx (10) and HPA (11). From GTEx we gathered RNA-Sequencing raw counts for all samples that were denoted with cause of death of traumatic injury, resulting in 421 samples from 42 tissues. From HPA we gathered RNA-sequencing reads for 192 tissues (ArrayExpress accession number: E-MTAB-2836). We converted these raw reads data into normalized counts as described elsewhere (39). Only genes with more than 10 normalized counts in at least one tissue were included in the analysis.
Protein–protein interactions data
Human PPIs were gathered from BioGrid (8), DIP (31), MINT (32) and IntAct (33) by using the MyProteinNet web-server (34). MyProteinNet ensures that only PPIs detected by established methods for physical interactions detection were considered.
Differential PPI scoring
Differential interactomes were created by using the following scheme: In each tissue, let 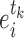 and
and 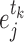 denote the normalized read counts of proteins
denote the normalized read counts of proteins 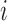 and
and 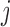 in tissue
in tissue 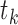 . Since the probability for an interaction between protein
. Since the probability for an interaction between protein 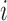 and protein
and protein 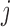 increases with their concentration, it can be approximated by
increases with their concentration, it can be approximated by 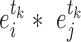 . Thus, we set the score of their interaction, denoted
. Thus, we set the score of their interaction, denoted 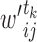 , to the sum of the log2 normalized read count values of proteins
, to the sum of the log2 normalized read count values of proteins 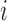 and
and 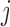 (Equation 1) in tissue
(Equation 1) in tissue 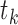 . We further normalized this score to fit the range of
. We further normalized this score to fit the range of 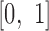 (Equation 2).
(Equation 2).
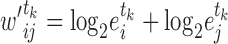 |
(1) |
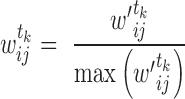 |
(2) |
After obtaining a score for each interaction in each tissue, the differential score of a specific interaction in tissue 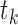 was computed by subtracting its median score across all tissues from its score in tissue
was computed by subtracting its median score across all tissues from its score in tissue 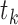 (Equation 3).
(Equation 3).
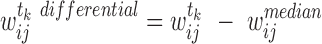 |
(3) |
Implementation
The DifferentialNet server was implemented in Python by using the Flask framework with data stored on a MySQL database. The website client was programmed using the ReactJS framework and designed with Semantic-UI. The network view is displayed by the cytoscape.js plugin (35). The website supports all major browsers. Recommended viewing resolution is 1440 × 900 and above.
Download
The DifferentialNet database is available for download under the permissive Creative Commons license. Downloadable data is versioned by numbered database builds and by global interactome build dates. The download page enables users to download data separately for each expression dataset.
Web-service access
The DifferentialNet database offers a web-service access to programmatically query the database by an expression dataset, tissue, threshold and genes. This is implemented via a REST-API method, which is callable via code or wget. More information can be found at http://netbio.bgu.ac.il/diffnet-api.
ACKNOWLEDGEMENTS
We thank Dr. Michal Gordon and Dr. Vered Chalifa-Caspi for their help in analyzing the HPA samples to obtain counts.
Footnotes
DifferentialNet: http://netbio.bgu.ac.il/diffnet
FUNDING
This work and the open access charge were funded by the Israel Science Foundation [860/13 to E.Y.-L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Schadt E.E., Friend S.H., Shaywitz D.A.. A network view of disease and compound screening. Nat. Rev. Drug Discov. 2009; 8:286–295. [DOI] [PubMed] [Google Scholar]
- 2. Vidal M., Cusick M.E., Barabasi A.-L.. Interactome networks and human disease. Cell. 2011; 144:986–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yeger-Lotem E., Sharan R.. Human protein interaction networks across tissues and diseases. Front. Genet. 2015; 6:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rolland T., Tasan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R. et al. A proteome-scale map of the human interactome network. Cell. 2014; 159:1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F. et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015; 163:712–723. [DOI] [PubMed] [Google Scholar]
- 6. Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K. et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015; 162:425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wan C., Borgeson B., Phanse S., Tu F., Drew K., Clark G., Xiong X., Kagan O., Kwan J., Bezginov A. et al. Panorama of ancient metazoan macromolecular complexes. Nature. 2015; 525:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N.K., O’Donnell L., Oster S., Theesfeld C., Sellam A. et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017; 45:D369–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Snider J., Kotlyar M., Saraon P., Yao Z., Jurisica I., Stagljar I.. Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 2015; 11:848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. GTEx Consortium The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015; 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 12. Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H. et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014; 509:582–587. [DOI] [PubMed] [Google Scholar]
- 13. Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. et al. A draft map of the human proteome. Nature. 2014; 509:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bossi A., Lehner B.. Tissue specificity and the human protein interaction network. Mol. Syst. Biol. 2009; 5:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emig D., Kacprowski T., Albrecht M.. Measuring and analyzing tissue specificity of human genes and protein complexes. EURASIP J. Bioinforma. Syst. Biol. 2011; 2011:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guan Y., Gorenshteyn D., Burmeister M., Wong A.K., Schimenti J.C., Handel M.A., Bult C.J., Hibbs M.A., Troyanskaya O.G.. Tissue-specific functional networks for prioritizing phenotype and disease genes. PLoS Comput. Biol. 2012; 8:e1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greene C.S., Krishnan A., Wong A.K., Ricciotti E., Zelaya R.A., Himmelstein D.S., Zhang R., Hartmann B.M., Zaslavsky E., Sealfon S.C. et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015; 47:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Magger O., Waldman Y.Y., Ruppin E., Sharan R.. Enhancing the prioritization of disease-causing genes through tissue specific protein interaction networks. PLoS Comput. Biol. 2012; 8:e1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barshir R., Basha O., Eluk A., Smoly I.Y., Lan A., Yeger-Lotem E.. The TissueNet database of human tissue protein–protein interactions. Nucleic Acids Res. 2013; 41:D841–D844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barshir R., Shwartz O., Smoly I.Y., Yeger-Lotem E.. Comparative analysis of human tissue interactomes reveals factors leading to tissue-specific manifestation of hereditary diseases. PLoS Comput. Biol. 2014; 10:e1003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lage K., Hansen N.T., Karlberg E.O., Eklund A.C., Roque F.S., Donahoe P.K., Szallasi Z., Jensen T.S., Brunak S.. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20870–20875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ostrow S.L., Barshir R., DeGregori J., Yeger-Lotem E., Hershberg R.. Cancer evolution is associated with pervasive positive selection on globally expressed genes. PLoS Genet. 2014; 10:e1004239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ideker T., Krogan N.J.. Differential network biology. Mol. Syst. Biol. 2012; 8:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bandyopadhyay S., Mehta M., Kuo D., Sung M.K., Chuang R., Jaehnig E.J., Bodenmiller B., Licon K., Copeland W., Shales M. et al. Rewiring of genetic networks in response to DNA damage. Science. 2010; 330:1385–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gill R., Datta S., Datta S.. A statistical framework for differential network analysis from microarray data. BMC Bioinformatics. 2010; 11:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Landeghem S, Van Parys T, Van Dubois M., Inzé D., Van De Peer Y.. Diffany: an ontology-driven framework to infer, visualise and analyse differential molecular networks. BMC Bioinformatics. 2016; 17:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gambardella G., Moretti M.N., De Cegli R., Cardone L., Peron A., Di Bernardo D.. Differential network analysis for the identification of condition-specific pathway activity and regulation. Bioinformatics. 2013; 29:1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goenawan I.H., Bryan K., Lynn D.J.. DyNet: visualization and analysis of dynamic molecular interaction networks. Bioinformatics. 2016; 32:2713–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ha M.J., Baladandayuthapani V., Do K.A.. DINGO: differential network analysis in genomics. Bioinformatics. 2014; 31:3413–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D.. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004; 32:D449–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ceol A., Chatr Aryamontri A., Licata L., Peluso D., Briganti L., Perfetto L., Castagnoli L., Cesareni G.. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010; 38:D532–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aranda B., Achuthan P., Alam-Faruque Y., Armean I., Bridge A., Derow C., Feuermann M., Ghanbarian A.T., Kerrien S., Khadake J. et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010; 38:D525–D531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Basha O., Flom D., Barshir R., Smoly I., Tirman S., Yeger-Lotem E.. MyProteinNet: Build up-to-date protein interaction networks for organisms, tissues and user-defined contexts. Nucleic Acids Res. 2015; 43:W258–W263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D.. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016; 32:309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A.. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an Online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015; 43:D789–D798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xin J., Mark A., Afrasiabi C., Tsueng G., Juchler M., Gopal N., Stupp G., Putman T., Ainscough B., Griffith O. et al. MyGene.info and MyVariant.info: gene and variant annotation query services. 2015; doi:10.1101/035667. [DOI] [PMC free article] [PubMed]
- 39. Basha O., Barshir R., Sharon M., Lerman E., Kirson B.F., Hekselman I., Yeger-Lotem E.. The TissueNet v.2 database: a quantitative view of protein–protein interactions across human tissues. Nucleic Acids Res. 2017; 45:D427–D431. [DOI] [PMC free article] [PubMed] [Google Scholar]