Abstract
Rare diseases affect over a hundred million people worldwide, most of these patients are not accurately diagnosed and effectively treated. The limited knowledge of rare diseases forms the biggest obstacle for improving their treatment. Detailed clinical phenotyping is considered as a keystone of deciphering genes and realizing the precision medicine for rare diseases. Here, we preset a standardized system for various types of rare diseases, called encyclopedia of Rare disease Annotations for Precision Medicine (eRAM). eRAM was built by text-mining nearly 10 million scientific publications and electronic medical records, and integrating various data in existing recognized databases (such as Unified Medical Language System (UMLS), Human Phenotype Ontology, Orphanet, OMIM, GWAS). eRAM systematically incorporates currently available data on clinical manifestations and molecular mechanisms of rare diseases and uncovers many novel associations among diseases. eRAM provides enriched annotations for 15 942 rare diseases, yielding 6147 human disease related phenotype terms, 31 661 mammalians phenotype terms, 10,202 symptoms from UMLS, 18 815 genes and 92 580 genotypes. eRAM can not only provide information about rare disease mechanism but also facilitate clinicians to make accurate diagnostic and therapeutic decisions towards rare diseases. eRAM can be freely accessed at http://www.unimd.org/eram/.
INTRODUCTION
Rare diseases are usually caused by genetic disorders and stay throughout a patient's entire life. Featuring low prevalence, a rare disease is defined to affect fewer than 1 in 1500 people in the United States, while fewer than 1 in 2000 people in Europe. As clinicians often fail to make a final diagnosis due to the lack of recognizable syndrome, precision medicine is commonly adopted to select optimal therapies based on a patient's genetic content.
Along with increasing public awareness of rare diseases, much effort has been devoted to relevant preclinical and clinical research. For example, next-generation sequencing (NGS) has been used to identify genes that cause rare diseases (including some novel phenotypes), which is accompanied by a parallel need for large-scale phenotypic annotations (1,2). The Human Phenotype Ontology (HPO) (3), which intends to realize large-scale computational analysis of the human phenome (a set of all phenotypes expressed by a species), contains ∼116 000 terms to describe individual phenotypic anomalies (4); however, the gene-to-phenotype association has been established for only a limited number of rare diseases. Most recently, industrialization of rare disease treatment development was proposed to drive down the treatment cost (5). This will need to centralize expertise and resources, which are based on various databases.
Under these circumstances, standardization of a disease-based phenotype system is in urgent need to integrate clinical phenotypes and symptoms, which is usually overlooked in existing databases. Over the last three years, we have extensively collected phenotypes and symptoms of rare diseases from published medical literatures and clinical data; standardized and classified extracted information via different patterns and approaches; provided enriched clinical and molecular annotations for most rare diseases; and finally generated a rare disease annotation system called Encyclopedia of Rare Disease Annotations for Precision Medicine (eRAM), This results in a valuable resource for researchers and clinicians to conduct studies and practice in rare diseases.
DISEASE DEFINITION
Disease unification and cross-linkages
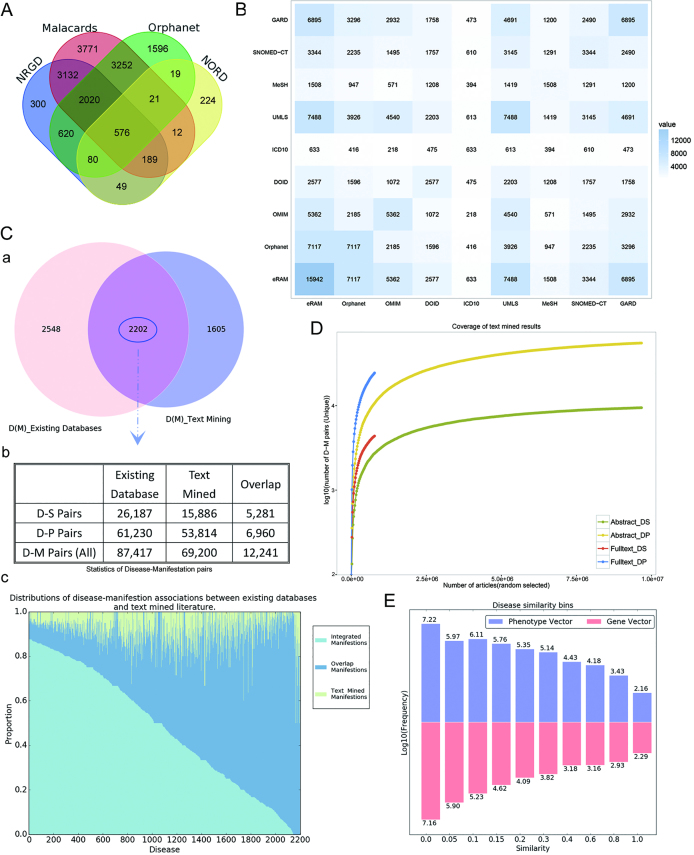
Currently, there is no unified, widely accepted definition for rare diseases, and rare diseases vary in prevalence throughout different populations (6). A need for collaboration across different countries has long been proposed to facilitate better definition, data sharing and diagnosis of rare diseases (7). In addition, considering the ubiquity of lexical heterogeneity in the realm of rare diseases, a well-structured, completed lexicon of rare diseases is necessary (8,9). To this end, we integrated data from four well-known databases – Orphanet (10), MalaCards (rare disease category) (9), NIH-Genetic and Rare Diseases (NGRD) (https://rarediseases.info.nih.gov/) and National Organization for Rare Disorders (NORD) (https://www.rarediseases.org) for rare diseases as disease name resources. We then mapped the disease names together with their alias strings to UMLS (2017AA release) through the lexical matching method (11,12) to complete the textual unification. Details together with an example of this method are provided in Supplementary 1. Given the fact that no existing standards/vocabularies can provide a complete list of standardized rare disease names, for those disease terms which cannot be mapped to UMLS, we adopted disease names from the Orphanet database as candidate vocabularies to standardize disease names since it defines each rare disease as a recognizable and homogeneous clinical presentation. Moreover, the Orphanet is widely accepted and used by clinicians and researchers (13). As a result, 14 771 unique disease concepts were obtained (Figure 1A). In addition, because of Orphanet's policy of unifying several Online Mendelian Inheritance in Man (OMIM) disease subtypes into one entry, to integrate and present rare diseases in a more accurate way, we then used OMIM to add disease subtypes as an expansion of rare diseases, through which we obtained the final disease concept list of 15 942 rare diseases. Considering the term usage variations in disease names and their identifiers (IDs), we mapped rare diseases among the currently controlled vocabularies and databases, including OMIM (14), Disease Ontology (DO) (15), ICD10, UMLS, Medical Subject Headings (MeSH), Systematized Nomenclature of Medicine - Clinical Terms (SNOMED-CT), GARD and Orphanet (Figure 1B). Different rare disease IDs mapped from the above databases were added as cross-linkage (Xref) annotations.
Figure 1.
(A) Overlaps among the major disease sources. Venn diagram for the major sources of disease names in eRAM. (B) Overlaps among all primary disease resources. The symmetric matrix shows the number of overlapping diseases between all pairs of primary name sources according to eRAM mapping. Colors and numerals represent the overlapping degree in disease counts. Source abbreviations: DOID – Disease Ontology (Identifier), GARD – NIH Rare Diseases. (C) (a) Overlap between diseases which have phenotypic annotations (phenotypes and symptoms) between existing databases and text-mining results. (b) Statistics of diseases and D-M pairs which have phenotypic annotations between existing databases and text-mined literature. (c) Distributions of disease-manifestation associations between existing databases and text-mined literature. The x-axis represents the diseases with phenotypic annotation from both existing databases and literature, while y-axis represents the proportion of disease corresponding manifestations. (D) Coverage of the text-mined result. Comparison of text-mined sentences containing phenotypes. The y-axis represents the number of sentences containing disease-related phenotypes for each disease. DS, Disease-Symptom pairs; DP, Disease-Phenotype pairs. (E) Distributions of disease similarity scores. Blue bins represent phenotypes (existing in 6147 diseases), and red bins represent genes (existing in 5593 diseases).
DISEASE ANNOTATION
After the disease unification process, we annotated rare diseases in the eRAM. Currently, each disease term in eRAM is annotated in eight aspects, including descriptions, synonyms, symptoms, genes, genotypes, Xref, human phenotypes and its relevant phenotypes in the mouse (MPO).
Disease descriptions
To help disambiguate the meaning through different disease terms, a definition/short description (if has) for each disease is provided. To maximize the description coverage for all the diseases in eRAM, we extracted 6322 disease definitions from MRDEF.RRF file in UMLS (2017AA), Orphanet, OMIM, DO, GARD and NORD. Up to now, 10 637 out of the 15 942 unique disease concepts in eRAM have their descriptions. Users can retrieve disease descriptions by clicking the ‘research’ button.
Disease symptoms and phenotypes
Accurate disease manifestations and sufficient clinical records are critical for the establishment of a rare disease annotation system. eRAM obtained symptom and phenotype information from the following sources: (i) human Phenotype Ontology (version 2017) (16). We extracted disease–phenotype (D–P) associations from HPO for all rare disease terms (including their synonyms) in eRAM. (ii) DO symptoms, using the ‘has symptom’ relationship. (iii) Orphanet. (iv) UMLS (2017AA) disease manifestation file—MRREL.RRF. As a result, a total of 16,944 phenotypes/symptoms were mapped to 1756 diseases. To ensure the high accuracy and integrity of the results, a pattern-based text mining approach was used to leverage external knowledge and limit the amount of human effort (17). To carry out this approach, we took the following two steps.
To build up a phenotype and symptom lexicon. The two most popular vocabularies containing disease manifestations are HPO and UMLS. Since the UMLS has now integrated the entire HPO (version 2017), we built up a comprehensive lexicon by extracting the symptom concepts as well as its synonym terms from UMLS (2017AA) symptoms using semantic type assignment of Sign or Symptom. Considering that the HPO has been not only adopted as a standard for phenotypic abnormalities but also treated as a computational bridge between genome biology and clinical medicine (18), thus allowing for deep phenotyping of rare diseases in health records and registries, we divided the whole lexicon into two sublexicons—the HPO terms as the ‘phenotype’ lexicon, and the UMLS unique manifestation terms as the ‘Symptom’ lexicon (19–21). As a result, 23 907 HPO terms (including alias strings) together with 16 178 UMLS unique manifestation terms (including alias strings) were obtained. The overlap terms between UMLS and HPO are 2212.
To develop the pattern dictionary. To develop the pattern dictionary that represents the relationship between disease and phenotype/symptom, we used the disease–manifestation (D–M) pairs in MRREL.RRF file from UMLS (2017AA) as the training source for disease-phenotype patterns and expanded both disease and phenotype/symptom concepts by mapping their corresponding synonyms from the whole UMLS (2017AA) Metathesaurus. Then, we extracted the syntactic patterns associated with the D–M pairs to train D–M specific patterns from abstracts and full-text articles in MEDLINE through a co-occurrence text mining approach. In total, 8 488 796 abstracts and 774 514 full-text articles were text-mined respectively from PubMed and PubMed Central, leading to the identification of 10 530 disease–symptom (D–S) pairs and 61 714 disease–phenotype pairs.
Next we applied the selected D-M patterns to text-mine the abstracts and full-text articles (from year 2010 to 2015) in MEDLINE using the pattern-based method. In total, 636 722 sentences together with 192 074 unique D–M (including alias strings) annotations were generated. The extracted D–M pairs were proved to be highly accurate (precision of 0.927, recall of 0.84 and F-score of 0.878) based on our manually selected 2000 pairs as test set (Supplementary 2). We then manually curated all the text-mined D-M results. Consequently, 181 978 out of 192 074 unique D–M pairs (including alias strings) were verified to be correct. The 181 978 D–M pairs are involved in 430 785 abstracts and 72 993 full-text articles. To evaluate the coverage of the text-mined D–M pairs, we calculated the number of unique D–M pairs extracted from articles in different size and observed the trend towards saturation. The extracted pairs from both abstracts and full-text articles showed a high coverage of phenotypic annotations of rare diseases. As expected, full-text articles contained more phenotypic and disease information than abstracts (Figure 1D). All the D–P and D–S sentences together with their PubMed identifiers (PMIDs) were retained for each rare disease.
Phenotype and symptom annotations in eRAM are represented separately. The annotations generated by HPO terms are shown in the ‘Phenotype’ tab, while the rest are shown in the ‘Symptom’ tab. All records consist of the results generated from text-mining and currently existing databases as previously mentioned (Figure 1C).
In addition, because of the wide application of animal models in better understanding human diseases, especially the mouse as the primary model organism in research on human biology and diseases, we mapped phenotype terms between HPO and Mammalian Phenotype Ontology (MPO) (22) based on both homologous gene mapping and lexical matching method. All information has been recorded in the eRAM.
Disease gene and genotype
The genotype refers to the genetic constitution of an individual, which is responsible for a particular trait. To better understand both etiology and mechanisms of disease, both gene and genotype information is necessary. In the present study, we collected disease-gene associations from several existing databases including Orphanet, OMIM, UniProtKB (23–25), ClinVar (26), DISEASES (including text-mined data) (27) and DisGeNET (CTD data) (28,29), as well as disease-gene associations inferred by the disease comorbidity-based network approach (30) using data in ClinVar. To make a better classification, we divided all these associations into three categories: curated (data manually curated by experts or validated by experiments), text-mined and inferred disease–gene associations. In total, eRAM contains 316 311 disease–gene association records currently, including 18 815 genes and 5593 diseases. For all genes, we collected, the corresponding locus information was also added. eRAM contains genotype information from the following resources: (i) existing databases: DisGeNET, GWASdb (31), LOVD (32) and PharmGKB (33); (ii) data from Beijing Children's hospital. In total, eRAM contains 92 580 gene variants. Users can view those data through clicking the ‘gene’ or ‘genotype’ button after querying a disease.
DISEASE CONNECTIONS
Connecting diseases with similar pathological mechanisms can inspire novel strategies on the effective repositioning of existing drugs and therapies (34). Usually, disease pairs sharing more involved genes or phenotypic information are more likely to have similar pathological mechanisms (35,36). Thus, we connected diseases by both phenotype-based and gene-based approaches (phenotypes and genes are curated or text-mined) using the following method.
Calculation of Phenotype-Based disease similarity
We adopted the equation of symptoms-based disease similarity introduced in previous work to calculate the phenotype-based disease similarity (37). The similarity ranges from 0 (no shared phenotype) to 1 (identical phenotypes). Details together with an example of phenotype-based disease similarity method are provided in Supplementary 3.
Calculation of Gene-Based disease similarity
We calculated gene-based disease similarity by determining the uniqueness of shared genes described in the former research (38). Details together with an example of gene-based disease similarity evaluation method are provided in Supplementary 4.
Users can retrieve top ten similar diseases based on phenotype-based or gene-based similarity for each disease from eRAM. The disease similarity generated from the phenotype-based method provides additional information on disease connections (Figure 1E), thus complementing the molecular biology-based classifying approach (20,39,40). In addition, to ensure the integrity of the gene-based disease connections, we combined disease-gene associations from all three categories, and obtained a gene-disease matrix connecting 17 324 genes with 5593 disease entries. The supplementary associations extracted from the inferred and text-mined categories intensify the disease network, suggesting much complicated relationships among diseases.
The gene-based and phenotype-based approaches expand connections among diseases in eRAM. However, when studying the mechanism-based disease connection in rare diseases, connections to common diseases are also very informative (41). Thus, we added connections between rare diseases and common diseases in two ways:
Connecting rare diseases to common diseases by gene based and phenotype-based methods. We integrated a common disease list by integrating common diseases in DO, and then we integrated disease–gene and disease–phenotype associations from HPO. In total, 9633 common diseases with 5317 disease-gene associations and 8906 disease-phenotype associations were generated.
Connecting rare diseases to common diseases by comorbidity. The comorbidity information in eRAM was mainly collected in two ways: (i) extracted from electronic health records (EHRs). We extracted the disease comorbidity information from Multiparameter Intelligent Monitoring in Intensive Care (MIMIC II) database, from which we collected 34 261 unique disease comorbidity pairs. This information has been presented in eRAM in the comorbidity section. (ii) Text-mined disease comorbidity information from MEDLINE. We first integrated disease concepts from eRAM, DO and OMIM, and obtained 171 938 disease terms (including synonyms). We then adopted the pattern-based approach described above to mine the literature from MEDLINE database. In total, 8 488 796 abstracts and 774 514 full-text articles were text-mined respectively, resulting in 142 422 unique disease comorbidity pairs with 356 845 sentences. The text-mining results were manually curated by experts in Beijing Children's hospital. All the text-mined sentences as well as their PMIDs have been deposited in eRAM.
DISCUSSION
Nowadays, rare diseases have drawn a lot of attention worldwide. NIH has launched Undiagnosed Disease Program for rare disease study and Canada has funded the Canadian FORGE (Finding of Rare Disease Genes) initiative (42). Similarly, the United Kingdom has conducted 100K genome project that includes a major focus on rare inherited diseases with the goal of introducing genomics diagnostics into the mainstream healthcare system for the benefit of patients and researchers (43). All of those projects rely on precisely defining the clinical phenotypes and symptoms. As a comprehensive platform for rare disease research and diagnoses, eRAM provides enriched clinical and molecular annotations for 15 942 rare diseases, consisting of integrated 6147 human disease-related phenotypic terms, 31 661 mammalian phenotypic terms, 10 202 symptoms standardized by UMLS, 18 815 genes and 92 580 genotypes, which provides systematic information combining clinical manifestations and molecular mechanisms. For convenient communication, a community-based disease annotation system has also been developed in the eRAM, where researchers and clinicians can exchange the latest advances and discoveries in rare diseases.
eRAM is delicate to providing rich and accurate knowledge that not only helps researchers to explore underlying mechanisms of rare diseases but also facilitates clinicians to make accurate diagnoses and therapeutic decisions. However, to develop a systematic and comprehensive database for rare diseases, more efforts remain to make. In the current eRAM, only 10,637 unique disease concepts have their corresponding descriptions. We will continuously collaborate with the experts from Beijing Children's Hospital and add as many short descriptions/summaries as possible for the rest 5305 diseases by integrating disease information from newly published articles and available clinical data. We will also continue to mine disease-manifestation associations from newly published abstracts/full-text articles. eRAM contains no performed phenotypic annotations extracted from the EHR database yet, mainly because the extracted information is in Chinese. In the future study, we will translate relevant annotations into English and integrate them into eRAM.
As the prevalence of rare diseases is extremely low, data sharing plays a critical role in exploring the diagnosis and mechanism of rare diseases. Thanks to Science China Life Sciences and Pediatric Investigation journals, we are authorized to host the related data about rare diseases published in these two journals. Under this policy, all de-identified clinical data with standardized phenotypes or manifestation terms will be deposited into eRAM. For example, all the relevant data of rare diseases published in the 2017 July special issue of Science China Life Sciences have been deposited into eRAM (44–55). In the future, we will continue to collect new rare disease cases, phenotypes and genotypes from published literature and other resources; meanwhile, we will standardize the electronic medical records for rare diseases from Beijing Children's Hospital and record those de-identified clinical data into eRAM. We plan to update annotations in eRAM every six months and change the version number every year.
The key point of precision medicine is to collect and analyze disease information from different individuals. To reach this goal, a well-structured and standardized database is needed to ensure the correct recording of patient-based data. With a rich accumulation of annotated phenotypes, clinical information, patient-based genotypes and phenotypes, eRAM will be the most comprehensive system to provide rare disease information, which is believed to facilitate the application of precision medicine for rare diseases in diagnosis and treatment selection. In the meantime, eRAM will serve as a useful source for exploring the underlying mechanism of rare diseases, while triggering the development of new therapeutic drugs.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Peng Li from Harvard Medical School for his constructive suggestions. We are also grateful to clinicians from Beijing Children's Hospital for their help with manual curation of the text-mined results.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
China Human Proteomics Project [2014DFB30010, 2014DFB30030]; National High Technology Research and Development Program of China [2015AA020108]; National Natural Science Foundation of China [31671377]; Shanghai 111 Project [B14019]; Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support [ZYLX201508]; Beijing Municipal Science and Technology Project [D131100005313014]. Funding for open access charge: China Human Proteomics Project [2014DFB30010, 2014DFB30030]; National High Technology Research and Development Program of China [2015AA020108]; National Natural Science Foundation of China [31671377]; Shanghai 111 Project [B14019]; Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support [ZYLX201508]; Beijing Municipal Science and Technology Project [D131100005313014].
Conflict of interest statement. None declared.
REFERENCES
- 1. Boycott K.M., Vanstone M.R., Bulman D.E., MacKenzie A.E.. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat. Rev. Genet. 2013; 14:681–691. [DOI] [PubMed] [Google Scholar]
- 2. Freimer N., Sabatti C.. The human phenome project. Nat. Genet. 2003; 34:15–21. [DOI] [PubMed] [Google Scholar]
- 3. Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S.. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008; 83:610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Groza T., Kohler S., Moldenhauer D., Vasilevsky N., Baynam G., Zemojtel T., Schriml L.M., Kibbe W.A., Schofield P.N., Beck T. et al. The human phenotype ontology: semantic unification of common and rare disease. Am. J. Hum. Genet. 2015; 97:111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ekins S. Industrializing rare disease therapy discovery and development. Nat. Biotechnol. 2017; 35:117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jia J., Shi T.. Towards efficiency in rare disease research: what is distinctive and important. Sci. China. Life Sci. 2017; 60:686–691. [DOI] [PubMed] [Google Scholar]
- 7. Mascalzoni D., Knoppers B.M., Ayme S., Macilotti M., Dawkins H., Woods S., Hansson M.G.. Rare diseases and now rare data?. Nat. Rev. Genet. 2013; 14:372. [DOI] [PubMed] [Google Scholar]
- 8. Trama A., Marcos-Gragera R., Sanchez Perez M.J., van der Zwan J.M., Ardanaz E., Bouchardy C., Melchor J.M., Martinez C., Capocaccia R., Vicentini M. et al. Data quality in rare cancers registration: the report of the RARECARE data quality study. Tumori. 2017; 103:22–32. [DOI] [PubMed] [Google Scholar]
- 9. Rappaport N., Twik M., Plaschkes I., Nudel R., Iny Stein T., Levitt J., Gershoni M., Morrey C.P., Safran M., Lancet D.. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017; 45:D877–D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pavan S., Rommel K., Mateo Marquina M.E., Hohn S., Lanneau V., Rath A.. Clinical practice guidelines for rare diseases: the orphanet database. PLoS One. 2017; 12:e0170365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fung K.W., McDonald C., Srinivasan S.. The UMLS-CORE project: a study of the problem list terminologies used in large healthcare institutions. J. Am. Med. Informatics Assoc: JAMIA. 2010; 17:675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCray A.T., Srinivasan S., Browne A.C.. Lexical methods for managing variation in biomedical terminologies. Proc. Symp. Comput. Appl. Med. Care. 1994; 235–239. [PMC free article] [PubMed] [Google Scholar]
- 13. Boycott K.M., Rath A., Chong J.X., Hartley T., Alkuraya F.S., Baynam G., Brookes A.J., Brudno M., Carracedo A., den Dunnen J.T. et al. International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. Am. J. Hum. Genet. 2017; 100:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A.. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015; 43:D789–D798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kibbe W.A., Arze C., Felix V., Mitraka E., Bolton E., Fu G., Mungall C.J., Binder J.X., Malone J., Vasant D. et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015; 43:D1071–D1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kohler S., Vasilevsky N.A., Engelstad M., Foster E., McMurry J., Ayme S., Baynam G., Bello S.M., Boerkoel C.F., Boycott K.M. et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res. 2017; 45:D865–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu R., Li L., Wang Q.. Towards building a disease-phenotype knowledge base: extracting disease-manifestation relationship from literature. Bioinformatics. 2013; 29:2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sifrim A., Popovic D., Tranchevent L.C., Ardeshirdavani A., Sakai R., Konings P., Vermeesch J.R., Aerts J., De Moor B., Moreau Y.. eXtasy: variant prioritization by genomic data fusion. Nat. Methods. 2013; 10:1083–1084. [DOI] [PubMed] [Google Scholar]
- 19. Soden S.E., Saunders C.J., Willig L.K., Farrow E.G., Smith L.D., Petrikin J.E., LePichon J.B., Miller N.A., Thiffault I., Dinwiddie D.L. et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci. Transl. Med. 2014; 6:265ra168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robinson P.N. Deep phenotyping for precision medicine. Hum. Mutat. 2012; 33:777–780. [DOI] [PubMed] [Google Scholar]
- 21. Kohler S., Schulz M.H., Krawitz P., Bauer S., Dolken S., Ott C.E., Mundlos C., Horn D., Mundlos S., Robinson P.N.. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 2009; 85:457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith C.L., Eppig J.T.. The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm. Genome. 2012; 23:653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. The UniProt, C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magrane M., UniProt C.. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011; 2011:bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boutet E., Lieberherr D., Tognolli M., Schneider M., Bansal P., Bridge A.J., Poux S., Bougueleret L., Xenarios I.. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol. Biol. 2016; 1374:23–54. [DOI] [PubMed] [Google Scholar]
- 26. Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pletscher-Frankild S., Palleja A., Tsafou K., Binder J.X., Jensen L.J.. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015; 74:83–89. [DOI] [PubMed] [Google Scholar]
- 28. Pinero J., Bravo A., Queralt-Rosinach N., Gutierrez-Sacristan A., Deu-Pons J., Centeno E., Garcia-Garcia J., Sanz F., Furlong L.I.. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017; 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pinero J., Queralt-Rosinach N., Bravo A., Deu-Pons J., Bauer-Mehren A., Baron M., Sanz F., Furlong L.I.. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database. 2015; 2015:bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu X., Jiang R., Zhang M.Q., Li S.. Network-based global inference of human disease genes. Mol. Syst. Biol. 2008; 4:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li M.J., Liu Z., Wang P., Wong M.P., Nelson M.R., Kocher J.P., Yeager M., Sham P.C., Chanock S.J., Xia Z. et al. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016; 44:D869–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pan M., Cong P., Wang Y., Lin C., Yuan Y., Dong J., Banerjee S., Zhang T., Chen Y., Zhang T. et al. Novel LOVD databases for hereditary breast cancer and colorectal cancer genes in the Chinese population. Hum. Mutat. 2011; 32:1335–1340. [DOI] [PubMed] [Google Scholar]
- 33. Thorn C.F., Klein T.E., Altman R.B.. PharmGKB: the pharmacogenomics knowledge base. Methods Mol. Biol. 2013; 1015:311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu C.C., Tseng Y.T., Li W., Wu C.Y., Mayzus I., Rzhetsky A., Sun F., Waterman M., Chen J.J., Chaudhary P.M. et al. DiseaseConnect: a comprehensive web server for mechanism-based disease-disease connections. Nucleic Acids Res. 2014; 42:W137–W146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pinero J., Berenstein A., Gonzalez-Perez A., Chernomoretz A., Furlong L.I.. Uncovering disease mechanisms through network biology in the era of Next Generation Sequencing. Scientific Rep. 2016; 6:24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nabhan A.R., Sarkar I.N.. Structural network analysis of biological networks for assessment of potential disease model organisms. J. Biomed. Inform. 2014; 47:178–191. [DOI] [PubMed] [Google Scholar]
- 37. Zhou X., Menche J., Barabasi A.L., Sharma A.. Human symptoms-disease network. Nat. Commun. 2014; 5:4212. [DOI] [PubMed] [Google Scholar]
- 38. Carson M.B., Liu C., Lu Y., Jia C., Lu H.. A disease similarity matrix based on the uniqueness of shared genes. BMC Med. Genet. 2017; 10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schofield P.N., Hancock J.M.. Integration of global resources for human genetic variation and disease. Hum. Mutat. 2012; 33:813–816. [DOI] [PubMed] [Google Scholar]
- 40. Griggs R.C., Batshaw M., Dunkle M., Gopal-Srivastava R., Kaye E., Krischer J., Nguyen T., Paulus K., Merkel P.A. Rare Diseases Clinical Research, N. . Clinical research for rare disease: opportunities, challenges, and solutions. Mol. Genet. Metab. 2009; 96:20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Z., Fang H., Slikker W., Tong W.. Potential reuse of oncology drugs in the treatment of rare diseases. Trends Pharmacol. Sci. 2016; 37:843–857. [DOI] [PubMed] [Google Scholar]
- 42. Beaulieu C.L., Majewski J., Schwartzentruber J., Samuels M.E., Fernandez B.A., Bernier F.P., Brudno M., Knoppers B., Marcadier J., Dyment D. et al. FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am. J. Hum. Genet. 2014; 94:809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McGrath J.A. Rare inherited skin diseases and the Genomics England 100 000 Genome Project. Br. J. Dermatol. 2016; 174:257–258. [DOI] [PubMed] [Google Scholar]
- 44. Ni X., Shi T.. The challenge and promise of rare disease diagnosis in China. Sci. China. Life Sci. 2017; 60:681–685. [DOI] [PubMed] [Google Scholar]
- 45. Wu D., Gong C., Su C.. Genome-wide analysis of differential DNA methylation in Silver-Russell syndrome. Sci. China. Life Sci. 2017; 60:692–699. [DOI] [PubMed] [Google Scholar]
- 46. Wang Y., Gong C., Wang X., Qin M.. AR mutations in 28 patients with androgen insensitivity syndrome (Prader grade 0–3). Sci. China. Life Sci. 2017; 60:700–706. [DOI] [PubMed] [Google Scholar]
- 47. Bai D., Shi W., Qi Z., Li W., Wei A., Cui Y., Li C., Li L.. Clinical feature and waveform in infantile nystagmus syndrome in children with FRMD7 gene mutations. Sci. China. Life Sci. 2017; 60:707–713. [DOI] [PubMed] [Google Scholar]
- 48. Cai S., Wang X., Zhao W., Fu L., Ma X., Peng X.. DICER1 mutations in twelve Chinese patients with pleuropulmonary blastoma. Sci. China. Life Sci. 2017; 60:714–720. [DOI] [PubMed] [Google Scholar]
- 49. Fu L., Jin Y., Jia C., Zhang J., Tai J., Li H., Chen F., Shi J., Guo Y., Ni X. et al. Detection of FOXO1 break-apart status by fluorescence in situ hybridization in atypical alveolar rhabdomyosarcoma. Sci. China. Life Sci. 2017; 60:721–728. [DOI] [PubMed] [Google Scholar]
- 50. Geng J., Wang H., Liu Y., Tai J., Jin Y., Zhang J., He L., Fu L., Qin H., Song Y. et al. Correlation between BRAF V600E mutation and clinicopathological features in pediatric papillary thyroid carcinoma. Sci. China. Life Sci. 2017; 60:729–738. [DOI] [PubMed] [Google Scholar]
- 51. Qi Z., Shen Y., Fu Q., Li W., Yang W., Xu W., Chu P., Zhang Y., Wang H.. Whole-exome sequencing identified compound heterozygous variants in MMKS in a Chinese pedigree with Bardet-Biedl syndrome. Sci. China. Life Sci. 2017; 60:739–745. [DOI] [PubMed] [Google Scholar]
- 52. Fang F., Liu Z., Fang H., Wu J., Shen D., Sun S., Ding C., Han T., Wu Y., Lv J. et al. The clinical and genetic characteristics in children with mitochondrial disease in China. Sci. China Life Sci. 2017; 60:746–757. [DOI] [PubMed] [Google Scholar]
- 53. Bai D., Zhao J., Li L., Gao J., Wang X.. Analysis of genotypes and phenotypes in Chinese children with tuberous sclerosis complex. Sci. China Life Sci. 2017; 60:763–771. [DOI] [PubMed] [Google Scholar]
- 54. Xu Z., Liu Y., Li H., Meng S., Boyd A.S., Stratton C.W., Ma L., Tang Y.W.. Detection of mycobacterial and viral DNA in Kikuchi-Fujimoto disease: an analysis of 153 Chinese pediatric cases. Sci. China Life Sci. 2017; 60:775–777. [DOI] [PubMed] [Google Scholar]
- 55. Li C., Zhang J., Li S., Han T., Kuang W., Zhou Y., Deng J., Tan X.. Gene mutations and clinical phenotypes in Chinese children with Blau syndrome. Sci. China Life Sci. 2017; 60:758–762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.