Abstract
There are a growing number of studies focusing on delineating genetic variations that are associated with complex human traits and diseases due to recent advances in next-generation sequencing technologies. However, identifying and prioritizing disease-associated causal variants relies on understanding the distribution of genetic variations within and among populations. The PGG.Population database documents 7122 genomes representing 356 global populations from 107 countries and provides essential information for researchers to understand human genomic diversity and genetic ancestry. These data and information can facilitate the design of research studies and the interpretation of results of both evolutionary and medical studies involving human populations. The database is carefully maintained and constantly updated when new data are available. We included miscellaneous functions and a user-friendly graphical interface for visualization of genomic diversity, population relationships (genetic affinity), ancestral makeup, footprints of natural selection, and population history etc. Moreover, PGG.Population provides a useful feature for users to analyze data and visualize results in a dynamic style via online illustration. The long-term ambition of the PGG.Population, together with the joint efforts from other researchers who contribute their data to our database, is to create a comprehensive depository of geographic and ethnic variation of human genome, as well as a platform bringing influence on future practitioners of medicine and clinical investigators. PGG.Population is available at https://www.pggpopulation.org.
INTRODUCTION
Recent advances in genotyping and sequencing technologies have facilitated genome-wide investigations on human genetic variations, as well as provided new insights into population history and genotype–phenotype relationships. The genetic background of an individual is a crucial factor that influences personalized medicine. It is now feasible to resequence an individual genome because of recent advances in next-generation sequencing (NGS) technologies. However, identifying and prioritizing disease-associated causal variants relies on understanding the distribution of genetic variation within and between populations. In this context, population genomics plays a vital role in dissecting the genetic architecture of complex traits/diseases by separating locus-specific effects from genome-wide effects, thereby serving as a bridge between evolution and medicine.
Over the past decades, many joint forces based on international collaborations have made remarkable achievements in studying human genetic variation, such as the Human Genome Diversity Project (1), the HapMap Project (2), the HUGO Pan-Asian SNP Project (3) and the 1000 Genomes Project (4). Nonetheless, these efforts have utilized highly heterogeneous ethnic groups, thus emphasizing the need for a more precise and comprehensive characterization of genomic diversity. In addition, the high mobility of human society in the recent decades has considerably increased the chances of interethnic marriages or genetic admixture, which in turn influenced genome diversity and further affected phenotypes relevant to health. Furthermore, the lack of adequate knowledge about the genetic structure of populations increases the risk of failure in sampling for complex traits/disease mapping (5).
Understanding prehistoric demographic events, such as population bottleneck, expansion, admixture, and natural selection not only facilitates insights into the extant pattern of genetic diversity of human populations but also plays essential roles in medical studies. First, it has fundamental implications for disease mapping. The population structure of target ethnic groups should be established prior to conducting association studies as it could be one of the major cofounding factors in the analysis (6). Genetic admixture detection enables a whole-genome admixture scan (admixture-mapping) to identify genetic factors underlying disease (7). Second, it is helpful for functional genomics. For instance, evolutionary conservation indicates functional importance. Detecting local adaptation signals enables us to study gene functions and how genes interact with the environment (e.g. the selection signals of EGLN1 and EPAS1 detected in Tibetan highlanders (8–10)). Third, demographic events could also alter the genetic load of a population (11–13), thus influencing medical genetics. Moreover, the genetic diversity shaped by demographic events may guide drug usage in cases involving variations in drug responses within and among populations (14).
It is helpful and necessary to dissect the genetic diversity and ancestral architectures for both evolutionary and genetic-based medical studies. To date, some databases have been built to curate the genetic relationships among different human groups using the Y chromosome and mtDNA (15). These databases have provided valuable resources for evolutionary genetics studies. However, so far few databases have focused on genomic diversity and genetic ancestry of human populations despite the accumulation of genomic data.
Here, we collected both genotyping and NGS data sets and integrated and re-analyzed the data to establish a special database, PGG.Population (www.pggpopulation.org), for understanding the genomic diversity and genetic ancestry of human populations. The first release of PGG.Population consists of 7122 genomes, covering 356 non-overlapping worldwide populations/groups. It presents a comprehensive description of the genomic diversity of each population, including their genetic affinity, population structure, genetic admixture, ancestral architecture, and footprints of natural selection in their genomes. Moreover, PGG.Population provides a user-friendly graphical interface and tools for users to analyze and visualize results in a dynamic style via online illustration. Our database is valuable to research groups that are interested in human genetic diversity, as well as medical and evolutionary history of ethnic groups in the context of population genomics. In particular, it facilitates both study design and results interpretation in studies of human populations.
MATERIALS AND METHODS
Data collection
We manually searched for information of each enrolled population online or from literatures (Figure 1 and Supplementary Table S1). Genome-wide genotyping data or NGS data of whole-genomes were collected for each human population. These genomic data covered not only general populations studied by international projects such as the HapMap Project (2), the Human Genome Diversity Project (1), the 1000 Genomes Project (4), the HUGO Pan-Asia SNP Project (3), the Human Origin data set (16) and the Simons Genomic Diversity Project (17), but also the indigenous/isolated populations contributed by regional sequencing efforts such as Tibetans (18), Sherpas (19), Xinjiang's Uyghurs (20) and ethnic groups with genomes deposited in Estonian Biocentre (http://evolbio.ut.ee/). The list of populations and genomes will be updated once new data are published. In the current version of PGG.Population, all the genome information were based on the Human Genome Build 37 positions. A full list of the data resources used can be found in Supplementary Table S1 (21–37).
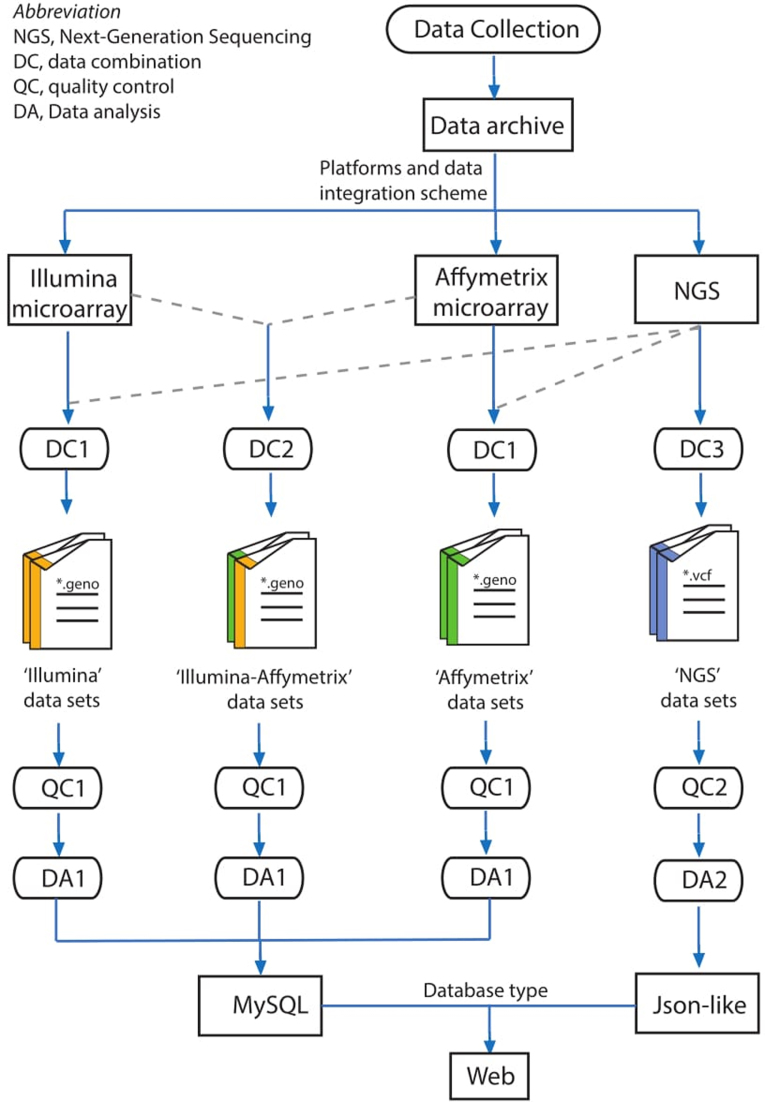
Figure 1.
An schematic diagram of data processing for PGG.Population database.
Data integration
Different genotyping data sets from diverse platforms were assembled for further analysis (Figure 1). For individual data genotyped on the same platform, such as Illumina arrays, these were directly pooled and then that of respective individuals were extracted with PLINK using filters that will be described in the next section (DC1 in Figure 1). In principle, we do not merge data obtained from distinct platforms (Illumina and Affymetrix arrays) owing to the small intersection of SNPs among them. However, combining data was applied when both the data of different platforms are valuable for understanding the demographic events of ethnic groups (DC2 in Figure 1). For example, we pooled individual data genotyped on Affymetrix Genome-wide Human SNP Array 6.0 and Illumina HO-Q Illumina HumanOmni1-Quad beadchip when exploring and reconstructing the population structure of Sherpas and Tibetans, of which the method was described in detail in Zhang et al. (19). For the purpose of minimizing strand ambiguity issues, all A/T and G/C markers were removed to reduce the risk of any ambiguity before data combination.
The NGS data were combined flexibly with (DC1 in Figure 1) or without genotyping data (DC3 in Figure 1), depending on the requirements of downstream analyses. When NGS data were combined with genotyping data, strand information was determined from the whole-genome sequence data based on the Human Genome Build 37 positions and strand was flipped to match that of the sequenced data. Only intersections of SNPs among NGS and genotyping data were retained for further analysis.
NGS data of high coverage (≥20×) were integrated from bam files for further analysis (DC3 in Figure 1). However, NGS data of low coverage (<20×) were not combined considering the VCF files were generated from data of different read depth, coverage and variant calling process. NGS reads were mapped to the human reference genome (Build 37) using Burrows-Wheeler Algorithm (38). SNP calling and raw variants filtering were carried out using the HaplotypeCaller module and the variant quality score recalibration (VQSR) module in GATK (39,40), respectively.
These steps thus generate different pooled data sets (‘Illumina’ data sets, ‘Affymetrix’ data sets, ‘Illumina-Affymetrix’ data sets, and ‘NGS’ data sets), which were flexible for reconstructing histories of diverse populations. We selected the latest and most representative data set for one group when the given population is covered by different datasets. A distinguished example is the Xinjiang's Uyghurs, where the data published by Feng et al. (20) were included as the best representative data set, as it consisted of around 1000 samples from diverse geographical regions.
Quality control
We filtered each combined data set that was assembled at both the single nucleotide polymorphism (SNP) and individual levels (QC1 in Figure 1). At the SNP level, we removed SNPs with call rate of <90% (across all individuals). At the individual level, we required at least 90% genotyping completeness for each individual (across all SNPs). We also removed recently related individuals by filtering one individual from all pairs when identity by descent (IBD) was >35%. All of the analyses were performed with PLINK v1.07 (41). Only biallelic variants were used for downstream analysis. Outliers were removed based on principal components analysis (PCA) for each data set. For each ‘NGS’ data set, only nucleotide sites passed universal filters were retained, as variant calling can be challenging in complex genomic regions (17) (QC2 in Figure 1).
To test the batch effect for each merged data set, PCA and FST-based analysis were performed (QC1 in Figure 1). Population samples from the same group and genotyped on different platforms were particularly used for examining any potential batch effects. These population samples are expected to show close genetic affinity (FST < 0.004) and cluster together in PCA plot (42) given there is no considerable batch effect. Data sets with significant batch effect were excluded from further analysis.
Analysis of genomic diversity and inference of genetic ancestry
Y chromosomal haplogroups were determined on the basis of key mutations commonly used for nomenclature of human paternal lineages. We developed an algorithm to search all possible combinations of the key mutations used for nomenclature from our sequence data to determine the fine-scale paternal haplogroup that was affiliated with each sample. mtDNA haplogroups were defined as described by Weissensteiner et al. (43). To estimate genetic affinities, FST between each pair of populations was calculated following Weir and Cockerham (44). To investigate fine-scale population structures, we performed a series of PCA using EIGENSOFT (45). We applied ADMIXTURE v1.30 (46) for unsupervised clustering analysis. Because the model in ADMIXTURE does not take linkage disequilibrium (LD) into consideration, we pruned each dataset using an r2 cutoff of 0.1 in each continuous window of 50 SNPs, and advanced by 10 SNPs (–indep-pairwise 50 10 0.1) with PLINK v1.07. We ran ADMIXTURE with random seeds for the datasets from K = 2 to K = 20 with default parameters (–cv = 5) in 10 replicates for each K for each data set. We used runs of homozygosity (ROH) to measure genetic diversity for each population. Natural selection analysis was performed only for NGS data sets using SelScan (47).
Website design and database back-end
PGG.Population is available at https://pggpopulation.org and requires no username and password. It has been tested in Google Chrome, Apple Safari, Mozilla Firefox, and IE8 browsers. The static web technology used included HTML5, CSS, and Bootstrap framework (http://getbootstrap.com/). To enhance user experience, JavaScript, jQuery and ECharts (https://ecomfe.github.io/echarts-doc/public/en/index.html) were implemented. The dynamic web was built using Java and Spring MVC framework (http://projects.spring.io/spring-framework/). All data were stored and managed using MySQL (https://www.mysql.com/). The data of natural selection signals were JSON-formatted which could be recognized and plotted by LocusZoom.js (http://locuszoom.sph.umich.edu/) in the front webpage. We receive email inquires and give response timely at pggadmin@picb.ac.cn, any suggestions on the website and database are welcome.
DATABASE CONTENT
PGG.Population Statistics
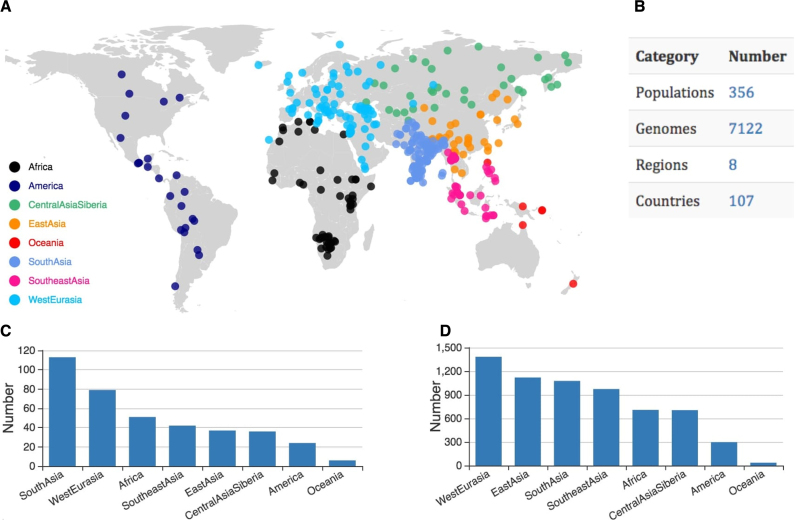
As of July 2017, PGG.Population consisted of 7122 genomes, representing 356 non-redundant populations/groups from 107 countries in 8 regions (Africa, America, Central Asia and Siberia, East Asia, Oceania, South Asia, Southeast Asia, and West Eurasia) collected from 27 studies (Figure 2 and Supplementary Table S1). Among these population entries, 49.1% are involved in ethnic groups from West Eurasia (n = 77) and South Asia (n = 98, among which 85 were collected from India). Despite of a vast of East Asian genomes (n = 1265) were included in our database, only a relative small number of populations from East Asia (n = 27) were represented, suggesting that previous studies from which we collected the data covered demographically large groups but ignored small/unique ones in this region. Our database covered the least number of Oceanian (5 populations and 36 genomes) and American (24 population and 297 genomes) samples at both the genome and ethnic group levels.
Figure 2.
Number of populations and genomes collected in the current release of PGG.Population. (A) Worldwide distribution of population samples, each belonging to one of the eight regional groups. (B) A summary of the population samples. (C) Number of groups in each region. (D) Number of genomes/individuals in each region.
Searching for population entries
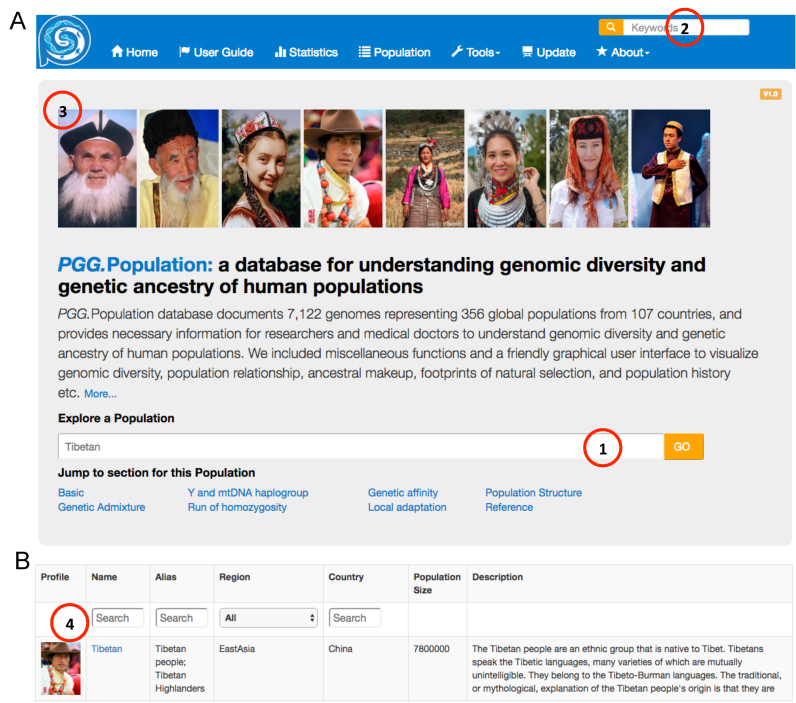
We provided user-friendly interfaces to query certain populations and explore their genomic diversity and evolutionary history. The first interface is keyword-based (Figure 3A). Users can build their query by inputting keywords related to ethnic group names. With keywords inputted by users, a SQL inquiry command is executed and a list of matched records appears in the front webpage in tabular form. If there is no matching record in our database, a ‘no result has been found’ page would appear and hints would be given for the queried task. The second interface is an advanced version (Figure 3B), in which users can select populations from given regions and countries or can input keywords for all items in the table that lists the populations. We also designed an interface that would randomly display eight population profiles every time one browses the welcome page (Figure 3A). Users can directly click one profile to access the webpage for the corresponding population.
Figure 3.
User-friendly interfaces for querying ethnic groups. (A) Keyword-based search bar. Users can find the bar in the welcome page of the database and in the upper right of any pages of our website. (B) Advanced interface to retrieving populations.
Population genomic diversity and genetic ancestry webpage
The genomic diversity and genetic ancestry page provides general information and comprehensive genetic analysis for a given population (for example, see https://www.pggpopulation.org/population/Tibetan?id=POP00001). The current release of PGG.Population contains the following eight sections that demonstrate different demographic events or information:
Basic information. A profile and brief introduction on the queried population is generated. The distribution(s) of the population (usually sampling locations) is illustrated on Google map to display the location and inhabited environment of the population.
Y chromosome and mtDNA haplogroups. Haplogroups of Y chromosome and mtDNA are informative genetic markers that may be utilized in forging human evolutionary contours. We used pie plots to show the proportion of each haplogroup observed in the population based on our database. We also estimated the probable ancestry to which the major haplotype belongs based on previous studies. When our database lacks of mtDNA and Y chromosome data for this population, then this section would be hidden. An interface/button has been included in this section to allow users to send us an email if they would like to contribute mtDNA and Y chromosome data.
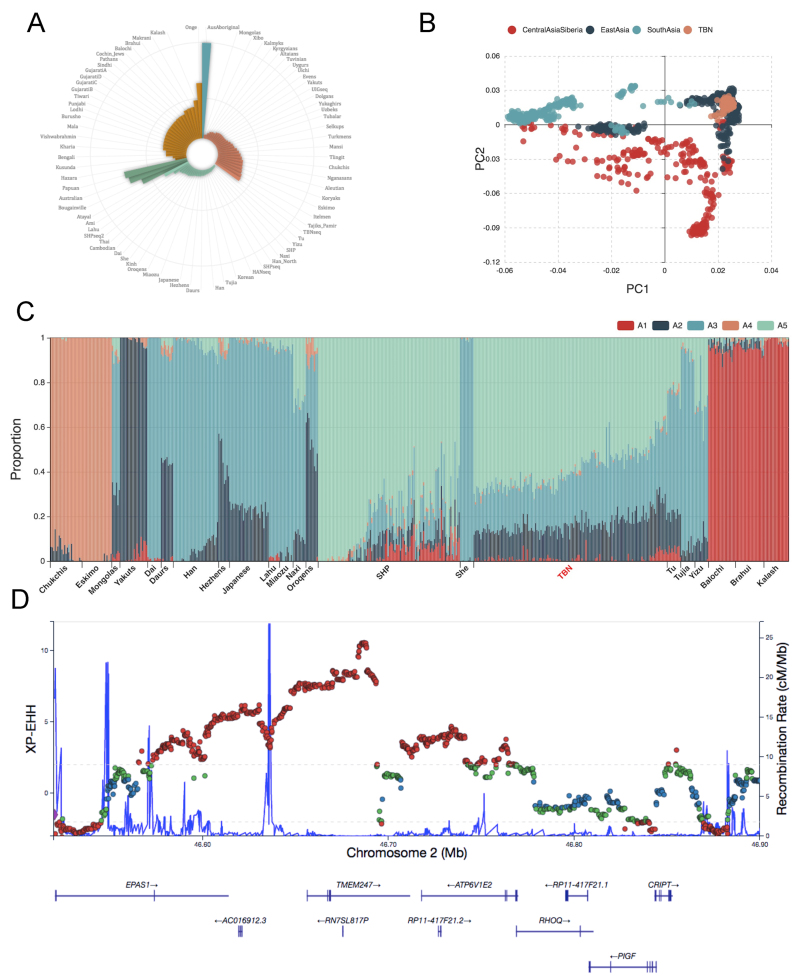
Genetic affinities. FST values of pairs of the quired population and other worldwide populations are calculated to determine their genetic relationship. For example, figure 4A shows that the overall genetic makeup of the Tibetan population is closest to Tu (FST = 0.012), Yizu (FST = 0.013), and Naxi (FST = 0.016) populations, followed by other surrounding East Asian populations and Central Asian/Siberian populations.
Population structure. PCAs were performed to investigate fine-scale population structures (Figure 4B). The structure of subgroups would also be displayed when necessary samples are offered. Examples include the sub-structures among Tibetan groups of culturally defined regions of historical Tibet (TBN.Qinghai, TBN.Chamdo, TBN.Lhasa, TBN.Nyingchi, TBN.Shanna, and TBN.Shigatse), Sherpas groups of China and Nepal (SHP.Zhangmu and SHP.Khumbu), and Uyghurs of Northeastern and Southeastern Xinjiang, China, which can be observed in the corresponding webpages.
ADMIXTURE. This section illustrates the result of an unsupervised clustering analysis, ADMIXTURE, based on which the user can investigate the genetic affinity and admixture pattern for one population (Figure 4C).
Runs of Homozygosity (ROH). ROH estimates genetic diversity of populations. If the overall ROH of one group is relatively lower compared to that of others, then isolation or inbreeding events may have occurred in this population. An example is the Sherpa people who historically underwent isolation (18).
Natural selection. In the current release of PGG.Population, iHS and XP-EHH were used to detect positive selection signals (Figure 4D). An interface button was included in this section that allows users to contact us if they would like to submit their NGS data to our database.
References. This section shows the references of the samples included in the genetic analysis of the webpage. All the genomes or samples can be traced to their resources.
Figure 4.
An example of genomic diversity and genetic ancestry reconstructed by PGG.Population for the Tibetan population. (A) A fan-like chart showing pairwise FST values between the Tibetan and other worldwide populations. The lengths of the bars are proportional to the FST values. (B) Plot of the first two principal components dissecting 22 East Asian populations. (C) Admixture analysis of 22 East Asian populations assuming five ancestral source populations. Each colour represents a genetic component. (D) An example of a genomic region showing signatures of natural selection.
Illustrating figures for users’ own analysis
We developed a web-based tool named ‘Figure Illustration’ to illustrate figures based on users' own analysis. The current version of the tool includes interfaces to plot PCA, ADMIXTURE and FST results. Users can directly upload their files that were generated by EIGENSOFT and ADMIXTURE to display their figures in a dynamic and interactive way. Moreover, users can use this tool to filter their data for quality control. For example, outliers in PCA could easily be identified and located by hovering the mouse over the element triggers, where an information box displays detailed information on a particular individual.
Interactive website elements
We provided several interactive website elements for each illustration item to enhance user experience. Here we use ADMIXTURE analysis on our website as an example to provide an introduction of the interactive events, and demonstrate how these can be manipulated to obtain the best solution scheme.
Mouse click event: Clicking on the color bars (or figure legends) will hide components represented by the corresponding color in the plot, and re-clicking on the inactive color (shown in grey) will enable it to show again on the plot.
Mouse hover event: Hovering on an element in a plot will trigger an information box containing detailed data on this element. For instance, by hovering a bar in the ADMIXTURE plot, users can find which population group one sample belongs to and the proportion of genetic ancestry for that individual.
Mouse wheel scroll: In ADMIXTURE and ROH plots, scrolling will zoom in/out the resolution of a specific plot, ranging from the minimum (1 individual) to the maximum (all samples).
Data view and figure download toolbox: For each plot, we prepared a toolbox for users to check numerical data and to download the adjusted figures. The buttons for these functions can be found upright in each plot, of which the page-shaped button will open a new box containing Tab-separated data, and the download button will convert the current plot into JPEG format as well as provide the download option.
Option menus: In the ADMIXTURE and ROH parts, we provided option menus where users could change the ancestry (K) numbers in the Admixture and the ROH length for descent. The options will change the level of analysis results and presentation.
Data download button. We provided a download button in each plot so that users can obtain the corresponding data underlying the figure.
FUTURE DIRECTIONS
The current version of PGG.Population documents 7122 genomes representing 356 global populations from diverse regions. We are aware that there are many ongoing sequencing efforts on indigenous ethnic groups particularly in Asia, where there is substantial genetic diversity. Therefore, extensive population data are expected to be included in the PGG.Population when available. The database will also extend the numbers of genomes and types of data for existing ethnic groups. Firstly, increasing sample size would provide unprecedented insights into the evolutionary history of a given population. For example, using ∼1000 of Xinjiang's Uyghur individuals and integrating worldwide samples, Feng et al. delineated the complex scenario of the admixture history of Uyghurs and revealed fine-scale population structures of Uyghurs (20). Secondly, there are different types of data/records of the human past. For instance, ancient DNA of modern humans and archaic genomes have been utilized in assessing human genomic diversity and reconstructing human histories (2). We are working on these data and will integrate these into the database in the near future. Furthermore, more information and extended analysis will be included in the database. The current version of PGG.Population provides basic information of the human populations enrolled and the most commonly used analysis of genomic diversity and genetic ancestry of human populations. In future versions, more comprehensive sets of evolutionary and population genetic parameters, such as effective population size, population mutation rates, recombination maps, gene flow, genetic load at both the population and individual levels, and adaptive genetic variants will be implemented. We will continuously improve the features and performance of each function to increase usage of the data and information in the database. Together with the joint efforts of other researchers who publish or republish their data in our database, the long-term ambition of the PGG.Population is to serve as a bridge between population genetic studies and future medical and clinical practices.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of Xu lab for their comments and suggestions on database features. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
The Strategic Priority Research Program [XDB13040100]; Key Research Program of Frontier Sciences [QYZDJ-SSW-SYS009] of the Chinese Academy of Sciences (CAS) (to S.X.); National Natural Science Foundation of China (NSFC) [91331204, 91731303, 31771388, 31711530221 to S.X.; 31501011 to Y.L.]; National Science Fund for Distinguished Young Scholars [31525014 to S.X.]; Program of Shanghai Academic Research Leader [16XD1404700 to S.X.]; National Key Research and Development Program [2016YFC0906403 to S.X.]; Science and Technology Commission of Shanghai Municipality (STCSM) [14YF1406800 to Y.L.]; S.X. is a Max-Planck Independent Research Group Leader and member of the CAS Youth Innovation Promotion Association; S.X. also gratefully acknowledges the support of the National Program for Top-Notch Young Innovative Talents of the ‘Wanren Jihua’ Project. Funding for open access charge: Key Research Program of Frontier Sciences of the Chinese Academy of Sciences [QYZDJ-SSW-SYS009].
Conflict of interest statement. None declared.
REFERENCES
- 1. Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L. et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008; 319:1100–1104. [DOI] [PubMed] [Google Scholar]
- 2. Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S.A., Harney E., Stewardson K., Fernandes D., Novak M. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015; 528:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hugo Pan-Asian SNP Consortium Mapping human genetic diversity in Asia. Science. 2009; 326:1541–1545. [DOI] [PubMed] [Google Scholar]
- 4. The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E.. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 2017; 100:635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N.. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008; 9:356–369. [DOI] [PubMed] [Google Scholar]
- 7. Smith M.W., O’Brien S.J.. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat. Rev. Genet. 2005; 6:623–632. [DOI] [PubMed] [Google Scholar]
- 8. Beall C.M., Cavalleri G.L., Deng L., Elston R.C., Gao Y., Knight J., Li C., Li J.C., Liang Y., McCormack M. et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z.X., Pool J.E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T.S. et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010; 329:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu S., Li S., Yang Y., Tan J., Lou H., Jin W., Yang L., Pan X., Wang J., Shen Y. et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol. Biol. Evol. 2011; 28:1003–1011. [DOI] [PubMed] [Google Scholar]
- 11. Simons Y.B., Turchin M.C., Pritchard J.K., Sella G.. The deleterious mutation load is insensitive to recent population history. Nat. Genet. 2014; 46:220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Henn B.M., Botigue L.R., Peischl S., Dupanloup I., Lipatov M., Maples B.K., Martin A.R., Musharoff S., Cann H., Snyder M.P. et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Henn B.M., Botigue L.R., Bustamante C.D., Clark A.G., Gravel S.. Estimating the mutation load in human genomes. Nat. Rev. Genet. 2015; 16:333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yasuda S.U., Zhang L., Huang S.M.. The role of ethnicity in variability in response to drugs: focus on clinical pharmacology studies. Clin. Pharmacol. Ther. 2008; 84:417–423. [DOI] [PubMed] [Google Scholar]
- 15. van Oven M., Kayser M.. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009; 30:E386–E394. [DOI] [PubMed] [Google Scholar]
- 16. Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D.. Ancient admixture in human history. Genetics. 2012; 192:1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A. et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016; 538:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu D., Lou H., Yuan K., Wang X., Wang Y., Zhang C., Lu Y., Yang X., Deng L., Zhou Y. et al. Ancestral origins and genetic history of Tibetan highlanders. Am. J. Hum. Genet. 2016; 99:580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang C., Lu Y., Feng Q., Wang X., Lou H., Liu J., Ning Z., Yuan K., Wang Y., Zhou Y. et al. Differentiated demographic histories and local adaptations between Sherpas and Tibetans. Genome Biol. 2017; 18:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feng Q., Lu Y., Ni X., Yuan K., Yang Y., Yang X., Liu C., Lou H., Ning Z., Wang Y. et al. Genetic history of Xinjiang's Uyghurs suggests Bronze Age multiple-way contacts in Eurasia. Mol. Biol. Evol. 2017; 34:2572–2582. [DOI] [PubMed] [Google Scholar]
- 21. Yunusbayev B., Metspalu M., Metspalu E., Valeev A., Litvinov S., Valiev R., Akhmetova V., Balanovska E., Balanovsky O., Turdikulova S. et al. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 2015; 11:e1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wong L.P., Lai J.K., Saw W.Y., Ong R.T., Cheng A.Y., Pillai N.E., Liu X., Xu W., Chen P., Foo J.N. et al. Insights into the genetic structure and diversity of 38 South Asian Indians from deep whole-genome sequencing. PLoS Genet. 2014; 10:e1004377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raghavan M., Skoglund P., Graf K.E., Metspalu M., Albrechtsen A., Moltke I., Rasmussen S., Stafford T.W. Jr, Orlando L., Metspalu E. et al. Upper palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014; 505:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., Kirsanow K., Sudmant P.H., Schraiber J.G., Castellano S., Lipson M. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014; 513:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kovacevic L., Tambets K., Ilumae A.M., Kushniarevich A., Yunusbayev B., Solnik A., Bego T., Primorac D., Skaro V., Leskovac A. et al. Standing at the gateway to Europe–the genetic structure of Western balkan populations based on autosomal and haploid markers. PLoS One. 2014; 9:e105090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wong L.P., Ong R.T., Poh W.T., Liu X., Chen P., Li R., Lam K.K., Pillai N.E., Sim K.S., Xu H. et al. Deep whole-genome sequencing of 100 southeast Asian Malays. Am. J. Hum. Genet. 2013; 92:52–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moorjani P., Thangaraj K., Patterson N., Lipson M., Loh P.R., Govindaraj P., Berger B., Reich D., Singh L.. Genetic evidence for recent population mixture in India. Am. J. Hum. Genet. 2013; 93:422–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fedorova S.A., Reidla M., Metspalu E., Metspalu M., Rootsi S., Tambets K., Trofimova N., Zhadanov S.I., Hooshiar Kashani B., Olivieri A. et al. Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of Northeast Eurasia. BMC Evol. Biol. 2013; 13:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Di Cristofaro J., Pennarun E., Mazieres S., Myres N.M., Lin A.A., Temori S.A., Metspalu M., Metspalu E., Witzel M., King R.J. et al. Afghan Hindu Kush: where Eurasian sub-continent gene flows converge. PLoS One. 2013; 8:e76748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yunusbayev B., Metspalu M., Jarve M., Kutuev I., Rootsi S., Metspalu E., Behar D.M., Varendi K., Sahakyan H., Khusainova R. et al. The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol. Biol. Evol. 2012; 29:359–365. [DOI] [PubMed] [Google Scholar]
- 31. Peng Y., Yang Z., Zhang H., Cui C., Qi X., Luo X., Tao X., Wu T., Ouzhuluobu, Basang et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol. Biol. Evol. 2011; 28:1075–1081. [DOI] [PubMed] [Google Scholar]
- 32. Metspalu M., Romero I.G., Yunusbayev B., Chaubey G., Mallick C.B., Hudjashov G., Nelis M., Magi R., Metspalu E., Remm M. et al. Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am. J. Hum. Genet. 2011; 89:731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chaubey G., Metspalu M., Choi Y., Magi R., Romero I.G., Soares P., van Oven M., Behar D.M., Rootsi S., Hudjashov G. et al. Population genetic structure in Indian Austroasiatic speakers: the role of landscape barriers and sex-specific admixture. Mol. Biol. Evol. 2011; 28:1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simonson T.S., Yang Y., Huff C.D., Yun H., Qin G., Witherspoon D.J., Bai Z., Lorenzo F.R., Xing J., Jorde L.B. et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010; 329:72–75. [DOI] [PubMed] [Google Scholar]
- 35. Rasmussen M., Li Y., Lindgreen S., Pedersen J.S., Albrechtsen A., Moltke I., Metspalu M., Metspalu E., Kivisild T., Gupta R. et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010; 463:757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Behar D.M., Yunusbayev B., Metspalu M., Metspalu E., Rosset S., Parik J., Rootsi S., Chaubey G., Kutuev I., Yudkovsky G. et al. The genome-wide structure of the Jewish people. Nature. 2010; 466:238–242. [DOI] [PubMed] [Google Scholar]
- 37. Reich D., Thangaraj K., Patterson N., Price A.L., Singh L.. Reconstructing Indian population history. Nature. 2009; 461:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H., Durbin R.. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011; 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. et al. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakatsuka N., Moorjani P., Rai N., Sarkar B., Tandon A., Patterson N., Bhavani G.S., Girisha K.M., Mustak M.S., Srinivasan S. et al. The promise of discovering population-specific disease-associated genes in South Asia. Nat. Genet. 2017; 49:1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weissensteiner H., Pacher D., Kloss-Brandstatter A., Forer L., Specht G., Bandelt H.J., Kronenberg F., Salas A., Schonherr S.. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016; 44:W58–W63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weir B.S. Estimating F-statistics: A historical view. Philos. Sci. 2012; 79:637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D.. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006; 38:904–909. [DOI] [PubMed] [Google Scholar]
- 46. Alexander D.H., Novembre J., Lange K.. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009; 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Szpiech Z.A., Hernandez R.D.. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 2014; 31:2824–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.