Abstract
Xenbase (www.xenbase.org) is an online resource for researchers utilizing Xenopus laevis and Xenopus tropicalis, and for biomedical scientists seeking access to data generated with these model systems. Content is aggregated from a variety of external resources and also generated by in-house curation of scientific literature and bioinformatic analyses. Over the past two years many new types of content have been added along with new tools and functionalities to reflect the impact of high-throughput sequencing. These include new genomes for both supported species (each with chromosome scale assemblies), new genome annotations, genome segmentation, dynamic and interactive visualization for RNA-Seq data, updated ChIP-Seq mapping, GO terms, protein interaction data, ORFeome support, and improved connectivity to other biomedical and bioinformatic resources.
INTRODUCTION
The African frogs, Xenopus laevis and X. tropicalis, have been widely used in biomedical research for many decades. As a tetrapod vertebrate, amphibians occupy a powerful phylogenetic niche between mammalian systems and less complex vertebrates. As a model system, the large and robust eggs and embryos of these frogs are extremely well suited to microinjection, microdissection, and transplantation based experiments, while their external development in a simple saline solution makes observation of responses to experimental intervention vastly more straightforward than are equivalent tests performed in internally developing species, such as mammals. The dual goals of Xenbase (www.xenbase.org) are to enhance and support research using these model organisms, and to make the wealth of unique data generated through the many advantages of this system available to the larger biomedical research community.
In the pre-genome era, Xenbase began with simple database modules supporting the Xenopus user community and papers describing research using Xenopus (1). As more powerful resources became available, new expressed sequence tags and draft genomes were added to build gene-centric features such as ‘Gene Pages’ and these data were linked to the literature via our Xenopus Anatomical Ontology, or XAO (2). With the addition of a manual data curation team in 2010 Xenbase expanded to manual annotation of literature published using Xenopus and began to capture gene expression and other content types such as morpholinos and antibodies and automatic systems that identify and link all of these elements (3). Most recently, we have added ORFeome, microRNA and human disease orthology content (4,5), and moved to a virtualized private cloud (6). In this paper, we introduce a number of major new features and enhancements including chromosome scale genome assemblies, a new genome browser, novel genome analysis tracks, a University of California, Santa Cruz (UCSC) Genome Browser (7) track hub and massive amounts of Chromatin Immunoprecipitation Sequencing (ChIP-Seq) and RNA sequencing (RNA-Seq) content. A novel RNA-Seq visualization system is also presented that allows users to load and view temporal, tissue, and experimental results from RNA-Seq data on-the-fly.
NEW X. laevis AND X. tropicalis GENOMES
An international multi-team effort for sequencing, assembly and annotation of the X. laevis genome was undertaken and took several years to complete (8). The genome sequence revealed that this species is an allotetraploid which evolved from a hybridization event between two species ∼17–18 million years ago. The two homeologous genes from each ancestor are still present for >56% of all genes, though the level of expression is often very different. As each X. laevis gene now has two copies in most cases, it was necessary to restructure both the user interface and the underlying database to seamlessly integrate the two homoeologs and to match each gene pair to the single corresponding gene in X. tropicalis (9). The chromosomes of one of the X. laevis ancestral species are longer than those of the second species. For this reason the Xenopus research community (10) chose to append gene symbols with a ‘.L’ (long) or a ‘.S’ (short) to indicate from which homoeologous chromosome they are derived, for example the pax8 homoeologs are pax8.L and pax8.S.
The X. laevis genome described by Session et al. (8) was released as version 9.1, and is viewable and searchable on Xenbase. The genome available at the National Center for Biotechnology Information (NCBI), known as version 9.2 GCF, has some minor differences to this version and is also available at Xenbase. A new chromosome-scale build of X. tropicalis was also released and is available for browsing, searching or download. The NCBI version of the X. tropicalis genome also differs from the genome consortium version, and is known as X. tropicalis NCBI v9.1. The official versions for future Xenbase annotation and Next Generation Sequencing (NGS) support are X. laevis version 9.2 GCF and X. tropicalis NCBI v9.1. Annotations for both of these builds were generated by Xenbase by merging the genome consortium and NCBI machine generated annotations using the BEDTools (11) intersect program. In the most recent version, 22,329 out of 51,041 gene models are annotated in X. laevis v9.2 GCF and 18,335 out of 34,207 gene models are annotated in X. tropicalis v9.1.
In addition to these latest genomes, Xenbase makes all previous genome versions available in genome browsers and source files accessible on its File Transfer Protocol (FTP) site (ftp://ftp.xenbase.org/pub/). Other resources also host some current and legacy Xenopus genomes, for details on these please see Vize and Zorn (12).
GENOME BROWSERS
For many years Xenbase has used GBrowse (13) both as a genome browser and to generate genome snapshots on Gene Pages. This browser uses the TopoView tool to stack High-throughput/NGS tracks—a powerful visualization methodology. In response to user requests, we also recently implemented a method to allow for Basic Local Alignment Search Tool (BLAST) searching from within GBrowse using a plugin written by Mark Wilkinson (https://github.com/GMOD/GBrowse). To use this tool, select the ‘Configure’ button in the top right corner of a GBrowse window, paste the sequence of interest into the text field, select ‘Configure’ again, and when back on the browsing window select ‘Go’. Note that running BLAST in this manner does not reload the genome view window, so if the matched region is not in the active view, it will not be visible. Users can always also go directly to our BLAST pages from the main navigation bar, and select the genome of interest as the target. The output will include hyperlinks to load and view the matched result directly in GBrowse.
As GBrowse is no longer supported by a developer community, the current Generic Model Organism Database (GMOD) community supported browser known as JBrowse (14) is being adopted as our primary genome browsing tool. In addition to our extant GBrowse resources, we now support JBrowse versions of the most recent genome builds and will focus on importing current and legacy genomes and tracks over to this new browser. JBrowse has many performance and interface improvements, and supports a powerful faceted track selector.
In addition to viewing and searching the genomes, we used GBrowse to generate snapshots of a genomic region to enrich Gene Pages and provide a graphical overview of the architecture of the gene of interest. JBrowse can render such views dynamically, but this method has a significant performance lag. As a step towards replacing GBrowse, we now use the data on gene coordinates and exon/intron boundaries already in our database to render a graphical representation of the genomic and chromosomal structure using Data-Driven Documents, JavaScript (D3.js; https://d3js.org/), a tool that will be discussed in more detail below. This provides instant rendering and allows for greater customization.
GENOME BROWSER TRACKS
In the past, Xenbase posted tracks submitted by genome sequencing centers and research groups that made their output available to our resource. More recently, we initiated in-house data analysis and track generation. The first set of custom genome-based data contains chromatin segmentation predictions and transcription factor binding site maps. Chromatin segmentation utilizes a combination of different histone modification markers and distribution of bound proteins such as p300 and RNA polymerase 2 to predict chromatin states and features such as promoters, enhancers, transcribed regions, repressed regions and heterochromatin, similar to the chromatin states described by Hontelez et al. (15). One aspect of Xenopus development that is intensively studied is how patterns of transcription change over time. To capture such transitions, Xenbase chromatin segmentation maps are generated from data derived from the same developmental time stage. Tracks are generated using ChromHMM (16) and Segway (17), similar to the UCSC genome browser. Our color scheme also matches that of the UCSC browser for consistency. Currently, this feature is available only for X. tropicalis NCBI version 9.1 as sufficient ChIP-Seq data are not available for X. laevis at this time.
Additional novel tracks include an 11-species conservation score track from an analysis by UCSC (unpublished) and a transcription factor binding site prediction track. Transcription factor binding site predictions were generated using motifs from the CIS-BP website (18) and scanning motif instances using FIMO/MEME (19). Between 20% and 70% of our predicted binding sites were validated by association with ChIP-Seq peaks, depending on the transcription factors.
An ongoing issue with genomic resources is data migration. Published NGS tracks are most often aligned to older genome builds, as the research/publication cycle moves slowly. In the past users needed to visit each legacy genome build and browse available tracks to view and evaluate all supporting content, and no single genome build had all available tracks. To alleviate this problem, Xenbase also rebuilds NGS tracks originally aligned against older genome builds to the latest builds, X. tropicalis NCBI v9.1 and X. laevis NCBI GCF v9.2. This includes RNA-Seq, ChIP-Seq, methylation and other NGS content. FASTQ files (20) obtained from NCBI are processed to generate BigWig (21) tracks for loading and viewing in JBrowse. This will make over 350 NGS tracks available on the latest genomes.
TRACK HUB
In addition to the GMOD browsers, we also support a UCSC browser track hub. This system uses the powerful resources at the UCSC to load and display content from the Xenbase remote site. Xenbase hosts a UCSC browser track hub, a collection of genomes and gene models plus over 350 NGS tracks, mostly ChIP- and RNA-Seq. All of this content can be loaded and displayed at UCSC, but tracks are only available against their original genome versions. As only the requested view is loaded at any one time, this system is very fast and has minimal load and display times. To access the hub visit Genomes/Xenbase UCSC Track Hub from the main navigation bar and follow the instructions. A second Xenopus data hub is available at Radboud University (http://veenstra.science.ru.nl/trackhub.htm).
INTERACTIVE CONTENT VISUALIZATION
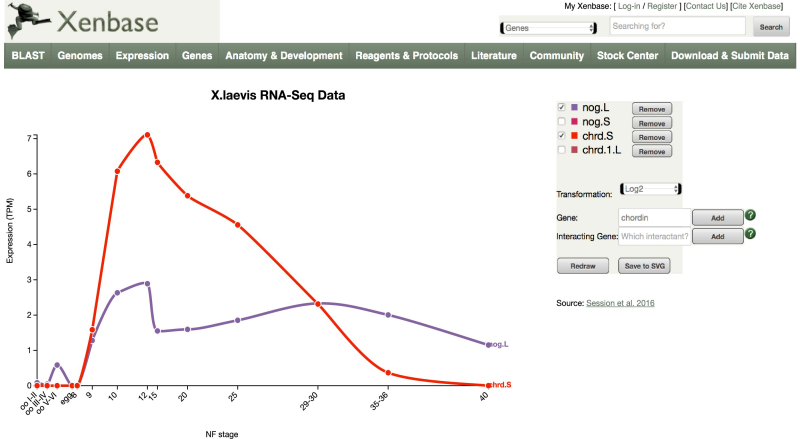
Databases typically display content generated through hard-coded queries and visualization methods in their web application code. Given the vast amount of NGS content available through resources such as NCBI GEO (22), and the enormous number of different potential unique uses by individual researchers, we developed a highly customizable method to view such data. The approach stores NGS data as a table in our core database. In response to a query, Xenbase pulls matching data from this table, and then formats the output as a simple JavaScript Object Notation (JSON) file that can be rendered and displayed by D3.js, a JavaScript library widely used in data visualization. The output graphic is dynamic and elements can be added or removed by the user. When adding new content that changes the scale, the visualization changes accordingly, and users can also select between different scaling methods for rendering the data values, such as non-transformed (normalized), log2 or square root. On each Gene Page, tools for viewing temporal and spatial expression data are now embedded in the Expression tab, and render as soon as the page is selected. Graph color is selected randomly, and reloading the Expression tab using the browser reload button, will select a new color combination. When a user clicks on one of the small icons, a larger version with new control tools opens, allowing new genes to be added, permanently removed, or temporarily hidden. Mouse-overs display the read counts for individual data points. The data used in this context are derived from the work of Session et al. (8) for X. laevis. For X. tropicalis genes, users have the option to view RNA-Seq data generated from different library protocols, or to view separate plots for different splice isoforms based on the data of Owens et al. (23).
In the example shown in Figure 1, the temporal profile of the noggin gene (nog) is compared to that of chordin (chrd), two genes identified by research in Xenopus as playing key roles in head development. The noggin profile was generated by navigating to the noggin gene page, selecting the Expression tab, and then clicking on the icon to view the ‘Stage and Tissue Profiles’ for X. laevis. This renders the X. laevis profile for both .L and .S homoeologs of noggin. The ‘Add’ feature was then selected, ‘chordin’ typed into the text field, and ‘Add’ clicked. The nog.S and chrd1.L genes were then removed to declutter the lower expressed homoeologs, and the transformation set to log2 to provide a more direct comparison. Users can also start from scratch by selecting Expression/RNA-Seq Viewers from the top horizontal navigation bar should they not wish to start with a Gene Page.
Figure 1.
Interactive RNA-Seq visualization. After selecting the central panel for X. laevis temporal data on a Gene Page ‘Expression’ tab, a larger window with additional controls opens as shown. A second gene, chordin, was added to further customize the view as described in the main text. Note that the noggin profile (purple) has been rescaled on the Y axis to accommodate the higher level of transcription of the chordin gene (red).
Tissue distribution data (8) of RNA-Seq data are also available via this tool as a heat map. This tool allows for simple comparison of spatial expression differences between the L and S homoeologs in X. laevis. Custom color maps, data transformations and gene comparisons will be available in the near future. Our data visualization interface also allows users to view changes in transcription of a gene in response to experimental manipulations, such as morpholino or mRNA microinjection, once again displayed as a heat map. This is available on Gene Pages in the Expression tab. Currently only one extensive dataset is available in this feature (24), but many more will be available in the near future.
All D3.js views can be output to a Scalable Vector Graphics (SVG) file, which can be edited in graphics programs (such as Adobe Illustrator) to generate figures for publications. The ‘Redraw’ button removes any custom text generated by mouse-overs to return to a clean rendering for download.
Xenbase is also in the process of building an RNA-Seq Transcripts Per Million (TPM) resource to enable users to rapidly evaluate and contrast datasets of interest. This involves annotating all available RNA-Seq datasets at the NCBI and building a prioritized queue for alignment and standardized normalization to output TPM tables. With further development, it is hoped that this content can be used to build global maps of how each gene behaves in all published Xenopus RNA-Seq datasets.
DATA IMPORTS FROM THE EUROPEAN BIOINFORMATICS INSTITUTE (EBI)
A number of data loaders now pull research data relevant to Xenopus from the EBI (25). This includes Gene Ontology (GO) terms for Xenopus proteins, Xenopus protein function descriptions, and human protein-protein interaction data. This content is updated monthly and included in our weekly data reports, available on our FTP site (ftp://ftp.xenbase.org/pub/GenePageReports/).
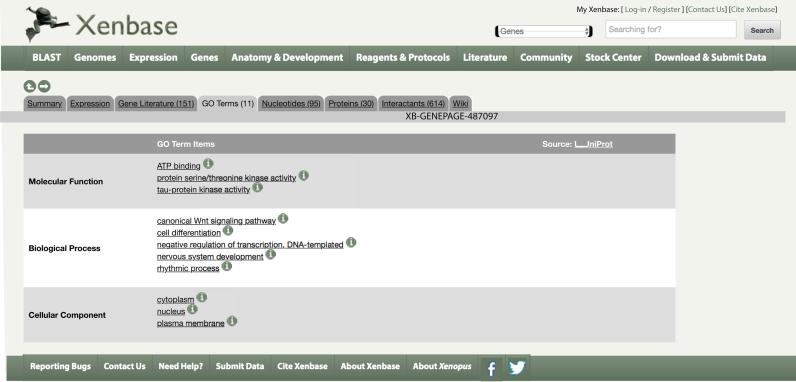
GO terms are collected for both X. laevis and X. tropicalis proteins. A non-redundant summary of these terms is available from the ‘GO Terms’ tab on every Gene Page as in Figure 2, and the full data, mapped to Xenbase identifiers (IDs), is placed on our FTP site on a weekly basis. On the Gene Page/GO Terms tab, the species and/or homoeolog to which the GO term is attributed can be viewed by mousing over the information icon. This new set of annotations is based on the UniProt GO annotation (UniProt-GOA) database, showing a filtered subset of manual and automated annotations meeting the criteria used by UniProtKB (25) for granularity, evidence code quality and excluding isoforms and annotations with certain qualifiers.
Figure 2.
GO terms associated with an X. laevis and X. tropicalis gene.
The UniProtKB also provides descriptions of protein function, along with the sources of the annotation. Xenbase now imports and displays this information near the top of the summary tab of each Gene Page as ‘Protein Function’. Click on the ‘+’ to expand if the description display is truncated.
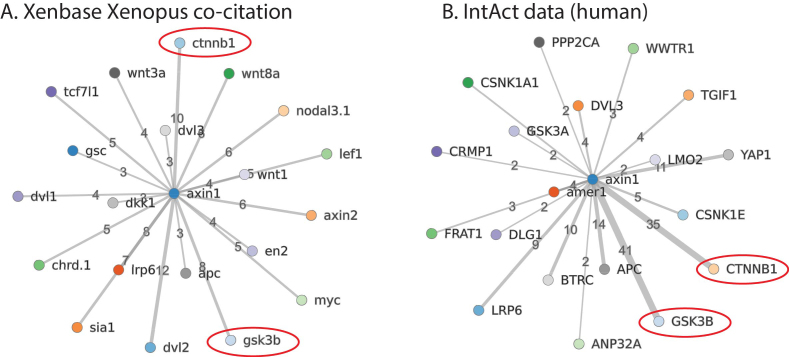
Xenbase now also imports protein-protein interaction content derived from human experimental data, updated monthly, from IntAct (26). This approach was selected as the amount of human data available is considerably larger than data currently available from research on Xenopus. We also display interactions predicted in-house from co-citation in the Xenopus literature. These are both available through the ‘Interactants’ tab on each Gene Page. D3.js is used to generate a dynamic view of the currently selected type of interactions, with edge thickness illustrating the number of evidence types or citations. Additional features will be added to allow further customization of the visualization by users, such as changing the number of root nodes. Figure 3 illustrates the top interactants for this new protein interaction data for AXIN1, compared to interactions predicted by co-citation of genes in the Xenbase literature. As predicted, there is often (but not always) considerable overlap in the two networks in terms of associated protein/gene identification, but the strength of evidence varies extensively.
Figure 3.
Comparison of Xenopus literature gene co-citation to human protein interaction data from IntAct. A. Genes co-cited with axin1 in Xenbase. Two key interactants, ctnnb1 (beta-catenin, 10 co-citations) and gsk3b (glycogen synthase kinase 3 beta, 8 co-citations) are circled in red. B. IntAct data on human proteins interacting with AXIN1. CTNNB1 has 35 results and GSK3B has 41 results, ranging from co-immunoprecipitation and yeast two-hybrid interactions, to fluorescence polarization spectroscopy and protein kinase assays (click on interactions hyperlink on Xenbase to view evidence).
ASSOCIATION WITH ADDITIONAL EXTERNAL RESOURCES
In addition to the already-existing human, mouse, and zebrafish links, Xenbase gene pages now have ortholog links to corresponding genes in fruit fly (27), chicken (28), and worm (29) MODs. The Genetic Phenotypes section has been augmented by links to OMIM (30), MalaCards (31), DECIPHER (32), KEGG (33) Disease and IMPC (34) mouse phenotype. In addition to matching OMIM IDs via gene symbols, Xenbase curators now also do manual annotation of Gene:OMIM associations from publications using Xenopus data. The Gene Expression section contains links to Ensembl (35) (multiple species), Protein Atlas (36), Eurexpress (37), Allen Brain Atlas for both human (38) and mouse (39) and MGI (40). Functional Ontologies now include AmiGO (41), PANTHER (42), and KEGG (33) Pathways links.
XenMine (43), a biological data mining tool set based on InterMine (44), has been available with Xenopus data since 2015. Links to XenMine are available from multiple places throughout the site, including gene pages.
ORFeome
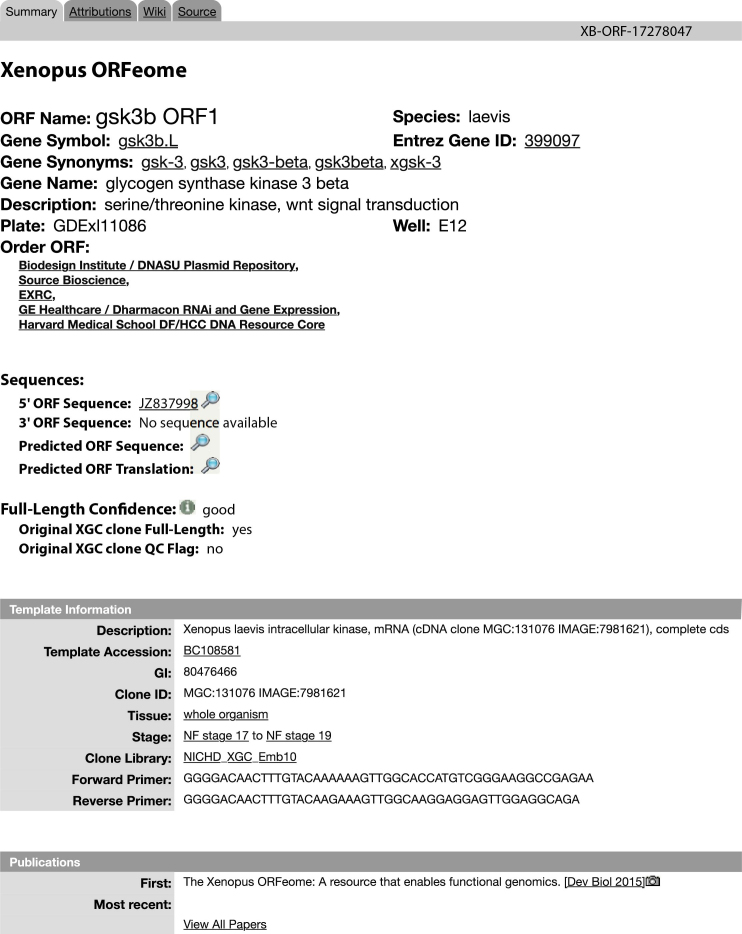
We now have full database support for the Xenopus ORFeome project (45). This project generated full length clones encoding the Open Reading Frame (ORF) of 8,633 Xenopus laevis cDNAs. These include 2,724 genes for which the human ortholog has an OMIM disease association. ORFeome cDNAs are cloned in Gateway vectors, and available from a number of commercial suppliers whose links are provided (they are not available directly from Xenbase). Users can search for ORFeome clones using ORF names or symbols, Entrez IDs, or plate and well numbers for specific clones. Links to ORFeome resources are also provided in the Reagents section of each Gene Page. The display for a typical ORFeome clone is shown in Figure 4. The ORFeome project is ongoing and additional clones, including those encoding X. tropicalis proteins, will be added as the data become available.
Figure 4.
ORFeome page associated with an X. laevis gene.
SMALL MOLECULES, microRNAs AND CRISPR/Cas
Three new wiki sections are available to support the use of small molecules, to provide data on microRNAs (miRNAs) and their expression, and a CRISPR/TALEN (46) support module. The small molecules wiki (main menu; Reagents & Protocols/Small Molecule Wiki) allows users to identify reagents that have been described in publications on Xenopus. Molecules are sorted by pathways (e.g. cAMP), by biological functions & processes (e.g. apoptosis), and by effects of classes of genes (e.g. ion channels). Each molecule has a wiki page that includes its description, known targets, suppliers, usage notes (if available), references, and molecular structure. Registered Xenbase users can submit content to the discussion section of each molecule page.
The miRNA wiki (main menu; Expression/miRNA Catalog) incorporates data and images from Ahmed et al. (47) with miRBase content. In addition, information on the sequence and mature product data images illustrating expression at multiple developmental stages are provided for each miRNA. More tools useful for miRNA work are supported via links to external resources, such as target prediction software, miRNA papers in Xenbase, and miRNA nomenclature.
We currently do not provide on-site CRISPR or TALEN target prediction or design tools. However, we do provide a summary and links to resources available at a number of specialized sites that do provide such services (main menu; Reagents & Protocols/ CRISPR & TALEN Support). This page summarizes which genome versions are available at each target prediction site, plus the sites input/output, aiding our users in selecting the best system for their experimental needs. It also provides links to various resources, publications and an educational genome editing video.
Xenbase HELP
Every Xenbase web page has a ‘Contact Us’ link in the top right corner that will lead to an email to report problems or ask for help. Throughout the resource, Information icons, a lower case ‘i’ in a green circle, provide additional data upon mouse-over. On the lower navigation bar on every page a ‘Need Help?’ button links to a suite of documentation on how to use various Xenbase features. Tutorial videos on using Xenbase are available at http://www.xenbase.org/other/static/HowTo.jsp. Finally, the Xenbase Wiki (wiki.xenbase.org) contains a wealth of helpful articles on Xenbase and Xenopus frogs and experimental protocols.
THE FUTURE
Our major targets for the immediate future are to expand and further improve the RNA-Seq visualization and analysis tools described above, and to add phenotype content including gene expression as a phenotype content. Numerous data-mining projects are also underway to identify synexpression sets within our extensive collection of gene expression data. We also have several active genome track improvement projects, mapping NGS data from old genomes to the latest genomes and generating ChIP-peak calls and other data to make NGS data more accessible. Finally, genome improvement and annotation is a major and high priority community need that we are addressing using a combination of computational and manual methods.
ACKNOWLEDGEMENTS
We would like to thank our NICHD program officer James Coulombe and our external advisory board for advice and guidance. Many members of our user community and external resources contributed greatly to the improvements described in this manuscript, including users who complete surveys and/or provide feedback, EBI/IntAct and Facebase. Research reported in this publication is supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
FUNDING
Research reported in this publication is supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development under Award Number P41HD064556. Additional support was provided by the Wellcome Trust via the EXRC [RRID:SCR_007164].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bowes J.B., Snyder K.A., Segerdell E., Gibb R., Jarabek C., Noumen E., Pollet N., Vize P.D.. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 2008; 36:D761–D767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Segerdell E., Bowes J.B., Pollet N., Vize P.D.. An ontology for Xenopus anatomy and development. BMC Dev. Biol. 2008; 8:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. James-Zorn C., Ponferrada V.G., Jarabek C.J., Burns K.A., Segerdell E.J., Lee J., Snyder K., Bhattacharyya B., Karpinka J.B., Fortriede J. et al. Xenbase: expansion and updates of the Xenopus model organism database. Nucleic Acids Res. 2013; 41:D865–D870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. James-Zorn C., Ponferrada V.G., Burns K.A., Fortriede J.D., Lotay V.S., Liu Y., Karpinka B.J., Karimi K., Zorn A.M., Vize P.D.. Xenbase: Core features, data acquisition, and data processing. Genesis. 2015; 53:486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karpinka J.B., Fortriede J.D., Burns K.A., James-Zorn C., Ponferrada V.G., Lee J., Karimi K., Zorn A.M., Vize P.D.. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res. 2015; 43:D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karimi K., Vize P.D.. The Virtual Xenbase: transitioning an online bioinformatics resource to a private cloud. Database (Oxford). 2014; 2014:bau108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tyner C., Barber G.P., Casper J., Clawson H., Diekhans M., Eisenhart C., Fischer C.M., Gibson D., Gonzalez J.N., Guruvadoo L. et al. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res. 2017; 45:D626–D634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Session A.M., UnoY. Kwon T., Chapman J.A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M. et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016; 538:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vize P.D., Liu Y., Karimi K.. Database and informatic challenges in representing both diploid and tetraploid Xenopus species in Xenbase. Cytogenet. Genome Res. 2015; 145:278–282. [DOI] [PubMed] [Google Scholar]
- 10. Matsuda Y., Uno Y., Kondo M., Gilchrist M.J., Zorn A.M., Rokhsar D.S., Schmid M., Taira M.. A new nomenclature of Xenopus laevis chromosomes based on the phylogenetic relationship to Silurana/Xenopus tropicalis. Cytogenet. Genome Res. 2015; 145:187–189. [DOI] [PubMed] [Google Scholar]
- 11. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vize P.D., Zorn A.M.. Xenopus genomic data and browser resources. Dev. Biol. 2017; 426:194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stein L.D., Mungall C., Shu S.Q., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.F., Harris T.W., Arva A. et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002; 12:1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H.. JBrowse: a next-generation genome browser. Genome Res. 2009; 19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hontelez S., van Kruijsbergen I., Georgiou G., van Heeringen S.J., Bogdanovic O., Lister R., Veenstra G.J.. Embryonic transcription is controlled by maternally defined chromatin state. Nat. Commun. 2015; 6:10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoffman M.M., Buske O.J., Wang J., Weng Z., Bilmes J., Noble W.S.. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods. 2012; 9:473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014; 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cock P.J.A., Fields C.J., Goto N., Heuer M.L., Rice P.M.. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010; 38:1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D.. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010; 26:2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Owens N.D., Blitz I.L., Lane M.A., Patrushev I., Overton J.D., Gilchrist M.J., Cho K.W., Khokha M.K.. Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome kinetics in development. Cell Rep. 2016; 14:632–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ding Y., Ploper D., Sosa E.A., Colozza G., Moriyama Y., Benitez M.D., Zhang K., Merkurjev D., De Robertis E.M.. Spemann organizer transcriptome induction by early beta-catenin, Wnt, Nodal, and Siamois signals in Xenopus laevis. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E3081–E3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. The UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004; 32:D452–D455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gramates L.S., Marygold S.J., dos Santos G., Urbano J.-M., Antonazzo G., Matthews B.B., Rey A.J., Tabone C.J., Crosby M.A., Emmert D.B. et al. FlyBase at 25: looking to the future. Nucleic Acids Res. 2017; 45:D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Antin P.B., Yatskievych T.A., Davey S., Darnell D.K.. GEISHA: an evolving gene expression resource for the chicken embryo. Nucleic Acids Res. 2014; 42:D933–D937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Howe K.L., Bolt B.J., Cain S., Chan J., Chen W.J., Davis P., Done J., Down T., Gao S., Grove C. et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016; 44:D774–D780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A.. OMIM.org: Online Mendelian Inheritance in Man (OMIM), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015; 43:D789–D798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rappaport N., Twik M., Nativ N., Stelzer G., Bahir I., Stein T.I., Safran M., Lancet D.. MalaCards: a comprehensive automatically-mined database of human diseases. Curr. Protoc. Bioinformatics. 2014; 47, doi:10.1002/0471250953.bi0124s47. [DOI] [PubMed] [Google Scholar]
- 32. Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P.. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 2009; 84:524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown S.D., Moore M.W.. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm. Genome. 2012; 23:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L. et al. Ensembl 2016. Nucleic Acids Res. 2016; 44:D710–D716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. et al. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 37. Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., Magen A., Canidio E., Pagani M., Peluso I. et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011; 9:e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012; 489:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oh S.W., Harris J.A., Ng L., Winslow B., Cain N., Mihalas S., Wang Q., Lau C., Kuan L., Henry A.M. et al. A mesoscale connectome of the mouse brain. Nature. 2014; 508:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blake J.A., Eppig J.T., Kadin J.A., Richardson J.E., Smith C.L., Bult C.J. the Mouse Genome Database Group . Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 2017; 45:D723–D729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S.. AmiGO Hub, Web Presence Working Group, AmiGO: online access to ontology and annotation data. Bioinformatics. 2009; 25:288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A.. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003; 13:2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reid C.D., Karra K., Chang J., Piskol R., Li Q., Li J.B., Cherry J.M., Baker J.C.. XenMine: a genomic interaction tool for the Xenopus community. Dev Biol. 2017; 426:155–164. [DOI] [PubMed] [Google Scholar]
- 44. Kalderimis A., Lyne R., Butano D., Contrino S., Lyne M., Heimbach J., Hu F., Smith R., Stěpán R., Sullivan J. et al. InterMine: extensive web services for modern biology. Nucleic Acids Res. 2014; 42:W468–W472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grant I.M., Balcha D., Hao T., Shen Y., Trivedi P., Patrushev I., Fortriede J.D., Karpinka J.B., Liu L., Zorn A.M. et al. The Xenopus ORFeome: a resource that enables functional genomics. Dev Biol. 2015; 408:345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nemudryi A.A., Valetdinova K.R., Medvedev S.P., Zakian S.M.. TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Naturae. 2014; 6:19–40. [PMC free article] [PubMed] [Google Scholar]
- 47. Ahmed A., Ward N.J., Moxon S., Lopez-Gomollon S., Viaut C., Tomlinson M.L., Patrushev I., Gilchrist M.J., Dalmay T., Dotlic D. et al. A database of microRNA expression patterns in Xenopus laevis. PLoS One. 2015; 10:e0138313. [DOI] [PMC free article] [PubMed] [Google Scholar]