Abstract
The microbiota is known to modulate the host response to influenza infection through as-yet-unclear mechanisms. We hypothesized that components of the microbiota exert effects through type I interferon (IFN), a hypothesis supported by analysis of influenza in a gain-of-function genetic mouse model. Here we show that a microbially associated metabolite, desaminotyrosine (DAT), protects from influenza through augmentation of type I IFN signaling and diminution of lung immunopathology. A specific human-associated gut microbe, Clostridium orbiscindens, produced DAT and rescued antibiotic-treated influenza-infected mice. DAT protected the host by priming the amplification loop of type I IFN signaling. These findings show that specific components of the enteric microbiota have distal effects on responses to lethal infections through modulation of type I IFN.
Significant heterogeneity in the host response to infection likely results from inherited as well as environmental factors. The enteric microbiota clearly interacts with the host to influence immune responses (1–5), possibly by integrating environmental signals. Influenza virus pathogenicity is highly influenced by the microbiota; infection of antibiotic-treated or germfree mice results in poor outcomes (6–8). How the microbiota exerts its protective effects remains incompletely understood, but we do know that microbial metabolites modulate a variety of important systemic phenotypes (5, 9–11). Possibly, specific microbial metabolites mediate protection against viral infection.
We focused on type I interferon (IFN) as an important signaling pathway in viral immunity in part because increasing evidence shows that the microbiota can regulate host immune homeostasis, as well as the response to injury and bacterial infection, through type I IFN signaling (12). However, its role during in vivo influenza infection remains uncertain, with multiple studies reporting dichotomous results (13–32). These studies primarily use loss of function of type I IFN signaling or administration of high levels of type I IFN. Given our hypothesis that microbially mediated changes in basal type I IFN levels influence outcomes in response to influenza, we used a genetic gain-of-function animal model, a mouse in which the immunity-related guanosine triphosphatase family M member 1 (Irgm1) is not expressed (Irgm1−/− knockout mouse). This model has a modest but functionally relevant threefold elevation in systemic type I IFNs compared with controls (33) and allows evaluation of the role of augmented type I IFN signaling before influenza infection.
To confirm that the lungs of Irgm1−/− mice expressed elevated type I IFNs, we used multiple assays. First, an IFN-specific varicella zoster virus infectivity bioassay of whole-lung tissue homogenate showed elevated type I IFN activity of Irgm1−/− lungs compared with littermate controls (Fig. 1A). Second, quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of type I IFN–stimulated genes (ISGs), Oas2 and Mx2, showed enrichment in Irgm1−/− lungs (Fig. 1B). Third, analysis of total RNA sequencing of control and Irgm1−/− lungs confirmed a robust signature for elevated type I IFN signaling (table S1). These assays support the utility of this model to study host responses to influenza infection in a background of elevated type I IFN in the lung.
Fig. 1. Irgm1−/− mice have elevated type I IFN in the lungs and are resistant to influenza.
(A) Bioassay for type I IFN activity (see Methods) from lung homogenates of littermate control and Irgm1−/− mice (n = 19 to 21 mice per group from five experiments). (B) Relative mRNA expression of Oas2 and Mx2 by qRT-PCR from control and Irgm1−/− lung homogenates (n = 7 to 14 mice per group from three to four experiments). (C) Kaplan-Meier survival analysis of infected control, Irgm1−/−, Ifnar−/−, and Irgm1−/−;Ifnar−/− mice (n = 18 to 40 mice per group from three to five experiments). (D) Viral load for control and Irgm1−/− mice infected with influenza (n = 11 to 23 mice per group from four to five experiments for days 3 and 6 and n = 4 to 7 mice from two experiments for day 10). (E) Viral RNA transcript counts in the lungs of infected control and Irgm1−/− mice at day 3 postinfection. (F and G) Representative images of lung cross sections from control and Irgm1−/− mice 6 days postinfection. Boxed areas are magnified immediately below. (F) Hematoxylin and eosin (H&E)–stained sections. Scale bar, 50 µm. (G) Sections stained for cleaved caspase 3 by immunohistochemistry. Scale bar, 20 µm. (H) Percentage of airways positive for at least one cleaved caspase-positive cell within an airway cross section [n = 5 to 6 mice per group from two experiments for (F) to (H)]. Means ± SEM. ND, not detected. *P ≤ 0.05; **P < 0.01; ****P < 0.0001; and ns, not statistically significant. Mann-Whitney used for statistical analysis in (A), (B), (D), (E), and (H). Mantel-Cox test was used with Bonferroni-corrected threshold in (C).
A dose of influenza that caused ~50% mortality of control mice showed minimal mortality and weight loss in Irgm1−/− littermates (Fig. 1C and fig. S1). However, influenza induced similar mortality and weight loss when these strains lacked the type I IFN receptor (IFNAR) (Fig. 1C and fig. S1). The lungs of Irgm1−/− mice had similar viral loads, kinetics, and viral antigen localization as controls (Fig. 1D and fig. S2). In contrast, viral transcripts were globally decreased in Irgm1−/− lungs compared with controls (Fig. 1E). Histopathological analysis of Irgm1−/− lungs compared with controls showed less airway epithelial damage with less apoptosis, a known mechanism of cell death during influenza infection (34, 35) (Fig. 1, F to H, and fig. S3). Analysis of RNA sequencing of lung tissue showed that infected controls had enhanced enrichment scores for proinflammatory innate immune responses and cell death pathways compared with infected Irgm1−/− lungs (table S2). As expected, infected Irgm1−/− lungs displayed reduced tissue levels of cytokines and chemokines previously associated with severe influenza in humans (36), including TNFα, MIP-1, interleukin-10 (IL-10), MCP-1, and interleukin receptor antagonist (IL-1Ra) (fig. S4).
The gut microbiota generates many small, diffusible metabolites that enter the systemic circulation (37). We hypothesized that specific metabolites protect from influenza pathogenesis by enhancing type I IFN signaling. Therefore, we screened the effects of 84 microbially associated metabolites (38) using a reporter cell line that harbors multiple type I IFN response elements (39). Each metabolite was screened for its induction of type I IFN signaling with polyinosinicpolycytidylic acid [poly(IC), a structural analog of double-stranded RNA] or amplification of type I IFN pathways with IFN-β treatment (fig. S5). We identified 11 metabolites that reproducibly increased reporter activity with either poly(IC) or type I IFN treatment (Fig. 2A). Three metabolites showed dose-dependent increases in reporter activity (Fig. 2B and fig. S6). Note that the level of activity was comparable to that found in high-throughput screens with this reporter for current drug discovery and design (39).
Fig. 2. A microbial-derived metabolite induces type I IFN activity.
(A) Scatter plots displaying fold change in luminescence for 100 µM metabolite screen in the presence of 5 mg/ml poly(IC) treatment (left) or 10 U/ml type I IFN treatment (right). (B) Fold increase compared with control in luminescence for DAT at the indicated doses in the presence of specified doses of poly(IC) or type I IFN (n = three to four experiments). (C) Stool and serum DAT levels measured by mass spectroscopy in mice treated with either vehicle (mock) or 2 weeks of antibiotics (n = 9 to 15 mice per group from three experiments). (D) Serum IFN activity as measured by the IFN bioassay of poly(IC)- and DAT-treated mice after 2 weeks of antibiotics (n = 7 or 8 mice per group from two experiments). αIfnar, antibody against IFNAR. (E) Relative mRNA expression of ISGs from lung homogenates of mice treated with or without DAT (n = 5 samples per group from two experiments). Means ± SEM. ND, not detected. *P < 0.05, **P < 0.01. Analysis of variance (ANOVA) used for statistical analysis in (D) (F = 10.9) with Sidak's multiple-comparisons test. Mann-Whitney test was used in (C) and (E).
One of these metabolites, desaminotyrosine (DAT), is potentially biologically relevant. DAT is a degradation product of flavonoids (40, 41), which comprise a group of polyphenolic compounds enriched in certain foods (42). Additionally, human enteric bacteria produce DAT from flavonoids and amino acid metabolism (40, 41, 43), and flavonoids have been proposed to exert immunoregulatory effects (44). In our facility, wild-type mice produce nanomoles of DAT per gram of feces and have picomolar quantities of DAT in the serum. By contrast, mice treated for 2 weeks with vancomycin, neomycin, ampicillin, and metronidazole (VNAM) had no fecal DAT and markedly reduced serum levels (Fig. 2C). Groups of mice were pretreated with VNAM followed by either 200 mM DAT or vehicle in drinking water for 1 week; concomitant treatment with oral DAT and systemic poly(IC) enhanced serum type I IFN activity (Fig. 2D). DAT treatment alone increased the expression of multiple ISGs in the lungs (Fig. 2E), potentially by augmenting production of type I IFN in response to ubiquitous viruses found in mice (45).
Groups of mice with or without VNAM pre-treatment were given 200 mM DAT or vehicle in drinking water 7 days before influenza infection and throughout infection for 14 days. Consistent with prior findings (6–8), antibiotic-treated mice showed increased mortality and weight loss compared with non–antibiotic-treated mice (Fig. 3A and figs. S7 and S8). Influenza-associated mortality and weight loss were less in DAT-treated mice compared with controls (Fig. 3A and figs. S7 to S9). Consistent with our hypothesis that DAT protects from influenza infection by enhancing type I IFN signaling before infection, DAT conferred no beneficial effect on weight loss or survival in Ifnar−/− animals (Fig. 3B and fig. S10). DAT protection was not H1N1 WSN strain–specific, as mice infected with PR8 and California/09 were protected by DAT (fig. S11).
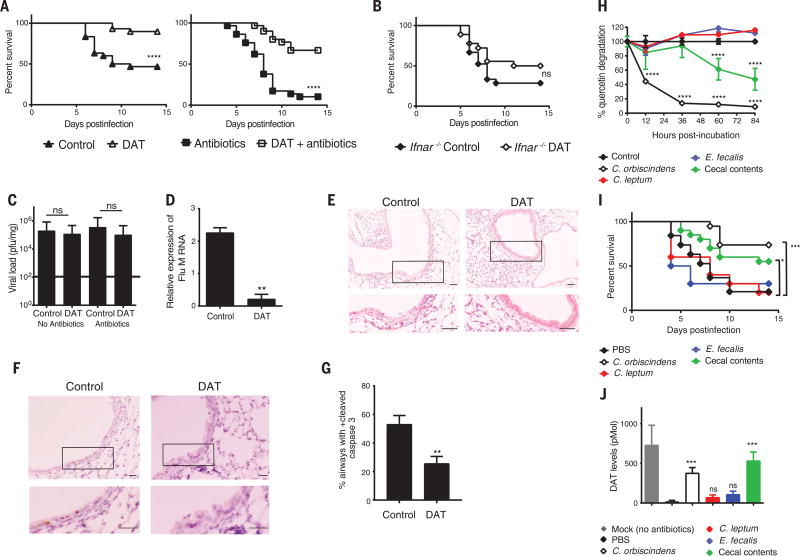
Fig. 3. DAT protects from influenza via type I IFN signaling.
(A) Kaplan-Meier survival analysis of groups of mice treated without antibiotics (left) or with a cocktail of broad-spectrum antibiotics (right, VNAM) and then treated with or without DAT before infection (n = 30 mice per group from two experiments, inclusive of all groups but plotted separately for clarity). (B) Kaplan-Meier survival analysis of Ifnar−/− mice treated with or without DAT and infected with influenza (n = 18 to 21 mice per group from two experiments). (C) Infectious viral load determined by plaque assay at day 5 postinfection for groups of mice from (A) (n = 10 mice per group from two experiments). (D) Matrix viral RNA expression determined by qRT-PCR from lung homogenates of mice treated with or without DAT as in (C). (E and F) Representative images of lung cross sections from mice treated with or without DAT at 5 days postinfection. Boxed areas are magnified immediately below. (E) H&E stained sections. Scale bar, 50 µm. (F) Sections stained for cleaved caspase 3 by immunohistochemistry. Scale bar, 20 µm. (G) Percentage of airways positive for at least one cleaved caspase–positive cell within an airway cross section [n = 10 mice per group from two experiments for (E) to (G)]. (H) Time course of quercetin degradation after incubation with control, mouse cecal contents, or single bacterial species (n = 6 replicates per group from two experiments). (I) Kaplan-Meier survival analysis of VNAM-pretreated mice gavaged twice with PBS, cecal contents, or single bacterial species and then infected with influenza (n = 20 mice per group from two experiments for PBS, cecal contents, C. orbiscindens gavage or 10 mice per group for C. leptum or E. fecalis gavage). (J) Stool DAT levels measured by mass spectroscopy at time of infection in mice from (I) (n = 10 per group). Statistical significance per group is compared with PBS-gavaged group. Means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; and ns, not statistically significant. Mantel-Cox test with or without Bonferroni-corrected threshold used in (A), (B), and (I). Mann-Whitney test used in (C), (D), and (G). ANOVA used in (H) (F = 50.8) and (J) (F = 14.5) with Dunnett's multiple-comparisons test.
We hypothesized that augmentation of type I IFN activity by DAT protects mice via a mechanism similar to that found in the Irgm1−/− mice. Indeed, DAT protection was not associated with a difference in viral titers 5 days postinfection (Fig. 3C), but we found less viral RNA in the lungs of mice treated with DAT than in controls (Fig. 3D). Similar to our findings in the Irgm1−/− gain-of-function model, greater airway epithelial damage and apoptosis were observed in control lungs than in DAT-treated mice (Fig. 3, E to G). These findings show that DAT also dampens host damage associated with influenza infection.
DAT generation occurs during flavonoid metabolism, and specific microbiota species metabolize flavonoids (46). A limited screen of the human fecal microbiota for flavonoid metabolism to DAT identified Clostridium orbiscindens (41) (Fig. 3H). Note that clostridial species are sensitive to metronidazole and vancomycin (47), the individual antibiotics that enhanced influenza-associated mortality (fig. S12). To investigate the role of specific bacteria in DAT generation and ultimately influenza protection, we obtained isolates of C. orbiscindens, C. leptum (a related bacterium), and Enterococcus faecalis (an unrelated microbiota member sensitive to vancomycin). Consistent with prior studies, we found that C. orbiscindens degraded flavonoid substrates effectively (Fig. 3H). Mouse cecal contents containing multiple prokaryote species also degraded the flavonoid substrate, albeit less effectively than pure cultures of C. orbiscindens (Fig. 3H). In contrast, other vancomycin-sensitive species, C. leptum and E. faecalis, did not degrade flavonoids (Fig. 3H). We gavaged groups of VNAM-pretreated mice with C. orbiscindens, mouse cecal contents, or phosphate-buffered saline (PBS) before influenza infection. C. orbiscindens and cecal content gavage protected mice from influenza mortality and morbidity (Fig. 3I and movies S1 to S3), whereas gavage with C. leptum or E. faecalis did not alter mortality, even though these organisms colonized mice efficiently (Fig. 3I and fig. S13). Moreover, C. orbiscindens and cecal contents restored fecal DAT levels, whereas C. leptum and E. faecalis did not (Fig. 3J).
Pretreating mice with DAT for 1 week before influenza infection, followed by cessation of treatment at the time of infection, protected mice from mortality and weight loss similarly to mice continuously treated (Fig. 4A and fig. S14). However, administration of DAT starting 2 days postinfection led to a worse outcome than for mice who never received DAT (Fig. 4A and fig. S14). This finding suggests that priming of the immune system by DAT before infection is protective. Consistent with this idea, Irgm1−/−;Rag1−/− mice, which maintain elevated levels of type I IFN, showed protection compared with Rag1−/− mice (fig. S15), and this implies that innate, not adaptive, immunity is crucial for the protective priming effect of type I IFN.
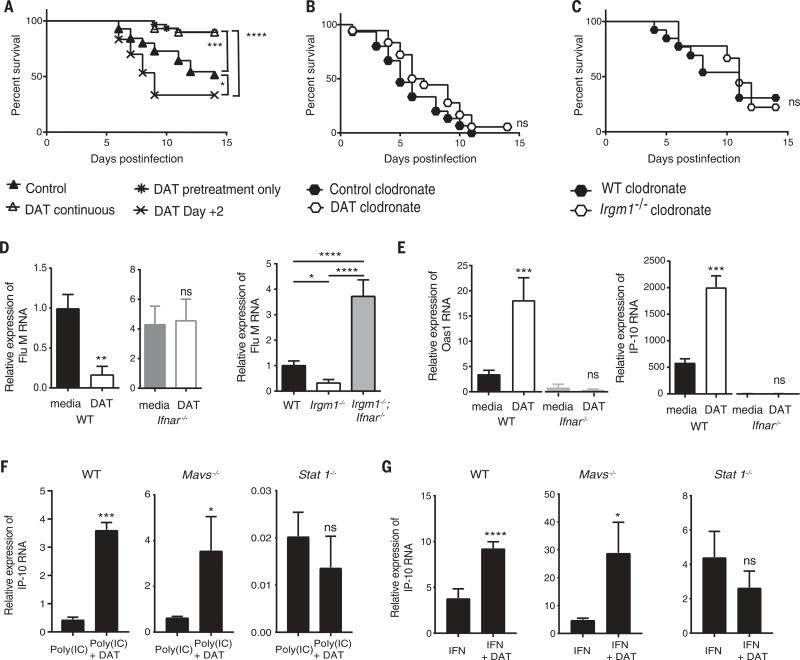
Fig. 4. DAT enhances type I IFN in macrophages via type I IFN amplification.
(A) Kaplan-Meier survival analysis of groups of mice with variations on the timing of DAT treatment with respect to infection. Control, no DAT; DAT continuous, 1 week pretreatment and continuous treatment postinfection; DAT pretreatment, 1 week pretreatment only; and DAT Day +2, treatment commenced 2 days postinfection (n = 30 mice per group from two experiments). (B) Kaplan-Meier survival analysis of mice all treated with clodronate liposomes and either control or DAT (n = 18 to 20 mice per group from two experiments). (C) Kaplan-Meier survival analysis of infected wild-type and Irgm1−/− mice all treated with clodronate liposomes (n = 9 to 13 mice per group from three experiments). (D) Relative expression of matrix influenza mRNA from wild-type and Ifnar−/− BMDMs pretreated with or without DAT and then infected (n = 7 replicates per group from two experiments for wild-type BMDMs and three replicates per group from three experiments for Ifnar−/− BMDMs). Matrix viral RNA expression determined by qRT-PCR from infected BMDMs from indicated genotypes (n = two to seven experiments). (E) Relative mRNA expression of Oas1 and IP-10 (as Cxcl10) by qRT-PCR from wild-type and Ifnar−/− BMDMs pretreated with or without DAT and then infected (n = 7 replicates per group from two experiments for wild-type BMDMs and three replicates per group from three experiments for Ifnar−/− BMDMs). (F) Relative mRNA expression of Cxcl10 (IP-10) by qRT-PCR from indicated genotypes treated with poly(IC) and with or without DAT (n = 4). (G) Relative mRNA expression of Cxcl10 (IP-10) by qRT-PCR from BMDMs from indicated genotypes treated with type I IFN and with or without DAT (n = 4). Graphs denote average with SEM displayed. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; and ns, not statistically significant. Mantel-Cox test used with or without Bonferroni-corrected threshold for statistical analysis in (A), (B), and (C). Mann-Whitney test used in (D) to (G). ANOVA in (D) (F = 37.5) used Tukey's multiple-comparisons test.
Within the lung, phagocytes are essential mediators of innate immune responses to bacterial and viral pathogens (48). To determine whether type I IFN mediates protection from influenza infection through a phagocytic-dependent mechanism, we treated mice systemically with clodronate liposomes to deplete lung phagocytes (49) (fig. S16). Clodronate treatment also abolished DAT- and Irgm1−/−-mediated protection (Fig. 4, B and C, and fig. S17). These results indicate that phagocytes are required for type I IFN–mediated protection from influenza infection.
Similarly to lung homogenates, influenza viral RNA was depleted in DAT-treated and Irgm1−/− bone-marrow–derived macrophages (BMDMs) in an Ifnar-dependent manner (Fig. 4D). DAT-pretreated BMDMs showed increased ISG transcripts after in vitro influenza infection, in an Ifnar-dependent manner (Fig. 4E). Similarly, ISG transcript abundance increased when DAT treatment was combined with either poly(IC) or type I IFN (Fig. 4, F and G).
Our initial validation of DAT did not discern between augmentation of type I IFN induction or type I IFN amplification. To clarify where DAT exerts its effects, BMDMs were isolated from mice genetically deficient in key mediators of type I IFN induction and amplification. To query the upstream induction pathway, we isolated BMDMs from Mavs−/− mice and found that DAT still enhanced poly(IC) and type I IFN expression of interferon-γ–induced protein 10 (IP-10) (Fig. 4, F and G). Therefore, we conclude that DAT is less likely to act in the induction pathway than in the amplification loop. To confirm this, we examined the role of STAT1, a signaling molecule downstream of IFNAR. IP-10 expression was not enhanced in similarly treated BMDMs from Stat1−/− mice (Fig. 4, F and G). This finding was confirmed with the STAT1 inhibitor, fludarabine (fig. S18). Taken together, these findings indicate that DAT augmentation of type I IFN signaling is mediated by IFN amplification via IFNAR and STAT1.
Our findings that preexisting members of the human microbiota protect the host from influenza infection may have implications for the known heterogeneous response to this infection in humans. Our results suggest that prior colonization by specific bacteria and a flavonoid-enriched diet are key components that modulate the immune response to influenza infection. Given that DAT offers a protective priming of the immune system, our findings also indicate the importance of timing in the ensuing overall immune response.
Supplementary Material
Acknowledgments
This work was supported by the Pediatric Scientist Development Program and funded by the National Institute of Child Health and Human Development, NIH 5K12HD000850-30 (A.L.S.), T32 DK007130-43 (G.P.C.), NIAID U19-AI070412 (MJH and TS), R01-AI111605 (MJH) and the Crohn’s and Colitis Foundation/Helmsley Charitable Trust. Additionally, we thank D. Kreamalmeyer for expertise in animal care and the Proteomics and Mass Spectrometry Facility at the Danforth Plant Science Center. All data to understand and assess the conclusions of this research are available in the main text and supplementary materials. RNA sequencing data are available via the following repository: ArrayExpress website, accession no. E-MTAB-5337. T.S.S., A.L.S., G.P.C., and G.E.K. are inventors on patent application no. 62413241 submitted by Washington University that covers the use of desaminotyrosine to enhance type I interferon stimulation.
Footnotes
References
- 1.Yang Y, et al. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu H, et al. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arpaia N, et al. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furusawa Y, et al. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 5.Smith PM, et al. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichinohe T, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abt MC, et al. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, et al. Nat. Commun. 2013;4:2106. doi: 10.1038/ncomms3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaiko GE, et al. Cell. 2016;165:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryu SH, Stappenbeck TS. Immunity. 2015;43:216–218. doi: 10.1016/j.immuni.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Trompette A, et al. Nat. Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 12.Kernbauer E, Ding Y, Cadwell K. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson S, Crotta S, McCabe TM, Wack A. Nat. Commun. 2014;5:3864. doi: 10.1038/ncomms4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koerner I, Kochs G, Kalinke U, Weiss S, Staeheli P. J. Virol. 2007;81:2025–2030. doi: 10.1128/JVI.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arimori Y, et al. Antiviral Res. 2013;99:230–237. doi: 10.1016/j.antiviral.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Högner K, et al. PLOS Pathog. 2013;9:e1003188. doi: 10.1371/journal.ppat.1003188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baskin CR, et al. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3455–3460. doi: 10.1073/pnas.0813234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beilharz MW, Cummins JM, Bennett AL. Biochem. Biophys. Res. Commun. 2007;355:740–744. doi: 10.1016/j.bbrc.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 19.Boon AC, et al. J. Virol. 2009;83:10417–10426. doi: 10.1128/JVI.00514-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung CY, et al. Lancet. 2002;360:1831–1837. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 21.Durbin JE, et al. J. Immunol. 2000;164:4220–4228. doi: 10.4049/jimmunol.164.8.4220. [DOI] [PubMed] [Google Scholar]
- 22.Fujikura D, et al. PLOS ONE. 2013;8:e55321. doi: 10.1371/journal.pone.0055321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Sastre A, et al. J. Virol. 1998;72:8550–8558. doi: 10.1128/jvi.72.11.8550-8558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobasa D, et al. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 25.Kugel D, et al. J. Virol. 2009;83:3843–3851. doi: 10.1128/JVI.02453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mordstein M, et al. PLOS Pathog. 2008;4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price GE, Gaszewska-Mastarlarz A, Moskophidis D. J. Virol. 2000;74:3996–4003. doi: 10.1128/jvi.74.9.3996-4003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szretter KJ, et al. J. Virol. 2009;83:5825–5834. doi: 10.1128/JVI.02144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tumpey TM, et al. J. Virol. 2007;81:10818–10821. doi: 10.1128/JVI.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng H, et al. J. Virol. 2007;81:12439–12449. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciancanelli MJ, et al. Science. 2015;348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oldstone MB, Teijaro JR, Walsh KB, Rosen H. Virology. 2013;435:92–101. doi: 10.1016/j.virol.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun L, et al. Cell Host Microbe. 2015;17:85–97. doi: 10.1016/j.chom.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takizawa T, et al. J. Gen. Virol. 1993;74:2347–2355. doi: 10.1099/0022-1317-74-11-2347. [DOI] [PubMed] [Google Scholar]
- 35.Mori I, et al. J. Gen. Virol. 1995;76:2869–2873. doi: 10.1099/0022-1317-76-11-2869. [DOI] [PubMed] [Google Scholar]
- 36.Gao R, et al. Am. J. Pathol. 2013;183:1258–1268. doi: 10.1016/j.ajpath.2013.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholson JK, et al. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto M, et al. Sci. Rep. 2012;2:233. doi: 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel DA, Patel AC, Nolan WC, Zhang Y, Holtzman MJ. PLOS ONE. 2012;7:e36594. doi: 10.1371/journal.pone.0036594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saura-Calixto F, et al. Mol. Nutr. Food Res. 2010;54:939–946. doi: 10.1002/mnfr.200900276. [DOI] [PubMed] [Google Scholar]
- 41.Schoefer L, Mohan R, Schwiertz A, Braune A, Blaut M. Appl. Environ. Microbiol. 2003;69:5849–5854. doi: 10.1128/AEM.69.10.5849-5854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chun OK, Chung SJ, Song WO. J. Nutr. 2007;137:1244–1252. doi: 10.1093/jn/137.5.1244. [DOI] [PubMed] [Google Scholar]
- 43.Lambert MA, Moss CW. J. Clin. Microbiol. 1980;12:291–293. doi: 10.1128/jcm.12.2.291-293.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Izzi V, et al. Front. Biosci. (Landmark Ed.) 2012;17:2396–2418. doi: 10.2741/4061. [DOI] [PubMed] [Google Scholar]
- 45.Virgin HW. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z, et al. PLOS ONE. 2014;9:e90531. doi: 10.1371/journal.pone.0090531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevens VW, et al. JAMA Intern. Med. 2017;177:546–553. doi: 10.1001/jamainternmed.2016.9045. [DOI] [PubMed] [Google Scholar]
- 48.Byrne AJ, Mathie SA, Gregory LG, Lloyd CM. Thorax. 2015;70:1189–1196. doi: 10.1136/thoraxjnl-2015-207020. [DOI] [PubMed] [Google Scholar]
- 49.van Rooijen N, van Nieuwmegen R. Cell Tissue Res. 1984;238:355–358. doi: 10.1007/BF00217308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.