Abstract
The renal biopsy provides critical diagnostic and prognostic information to clinicians including in cases of acute kidney injury, chronic kidney disease and allograft dysfunction. Today, biopsies are read using a combination of light microscopy, electron microscopy and indirect immunofluorescence with a limited number of antibodies. These techniques were all perfected decades ago with only incremental changes since then. By contrast, recent advances in single cell genomics are transforming scientists ability to characterize cells. Rather than measure expression of several genes at a time by immunofluorescence, it is now possible to measure expression of thousands of genes in thousands of single cells simultaneously. Here, we argue that the development of single cell RNA-sequencing (scRNA-seq) offers an opportunity to comprehensively describe human kidney disease at a cellular level. It is particularly well suited for the analysis of immune cells which are characterized by multiple subtypes and changing functions depending on their environment. In this review, we summarize the development of scRNA-seq methodologies. We discuss how these approaches are being applied in other organs, and the potential for this powerful technology to transform our understanding of kidney disease once applied to the renal biopsy.
Keywords: RNA sequencing, biopsy, informatics, microfluidics
Introduction
The kidney is a highly specialized organ consisting of almost one million nephrons per adult human kidney.1 Each nephron is a single functioning unit composed of a filtering unit, a glomerulus at one end followed by a continuous tubule that connects to a collecting duct that in turn delivers urine to the renal pelvis. The tubule of the nephron is divided into segments defined by unique physiologic role, location, ultrastructure and histologic feature. Each nephron is surrounded by a network of capillaries arranged in a unique way so as to allow the nephron segments reabsorb, excrete and concentrate filtrate among other functions. The nephron is surrounded by important supporting cells required for the maintenance of this complex system and performance of all the functions a normal kidney requires. All these specialized cell types perform a unique role and can exist in various states of activity and metabolism at any one time.
Understanding kidney function requires knowledge of the role that each individual cell type plays in this complex system. Defining the gene regulatory mechanisms that underlie cell behavior represents a question of fundamental importance in nephrology. This knowledge is key to the understanding of cellular function. Traditional approaches for characterization of kidney cell types have relied on visualization of tissue architecture using methods such as microscopy and fluorescence-activated cell sorting (FACS). These approaches have high spatial resolution but rely on a limited number of markers, precluding comprehensive characterization of kidney cell types and states. Furthermore, these methods are poor at revealing systematic gene expression changes occurring in individual cells.
The genome gives rise to the remarkable diversity of cell types through differences in gene expression. For this reason, profiling the genes actively transcribed in each cell is a powerful approach to categorizing the heterogeneous cell population found in the kidney. Over the last two decades, knowledge regarding the transcriptional profiles of cells in the kidney has come from whole organ profiling using either microarray2 or next generation RNA sequencing (RNA-seq).3, 4 In these experiments whole tissue blocks or sections are studied without regard for the heterogeneity of the sample, otherwise known as bulk sequencing. These studies have been highly informative, but are fundamentally limited to describing the averaged gene expression across a cell population. This approach may hide or skew signals of interest such as genes expressed in low abundance cell types or genes that may have an important effect even at low expression levels (such as transcription factors). Alternative approaches have attempted to achieve finer separation of kidney compartments to address the issue of mixed cell type signatures. For example, Lee et al microdissected 14 tubule segments and analyzed the gene expression of each individual segment.5 But this approach does not reveal individual cell states, and cannot distinguish separate cell types within a particular segment such as the principal and intercalated cells. Laser capture microdissection can achieve compartment-specific transcriptional profiles, but like microdissection, it cannot resolve interstitial or glomerular cell types.6
Other more recent advances have improved the ability to study gene expression from individual cells. Two examples are RNA-seq of FACS sorted cells7 and translating ribosome affinity purification (TRAP)8–10. These approaches have provided great insight into the molecular signatures and gene regulatory networks for specific cell types in kidney development, homeostasis and disease. The former method can isolate a population of cells from tissue defined by known cell type markers. The latter technique allows one to focus on genes that are actively being transcribed in a genetically defined cell type of interest. However, these techniques require advance knowledge of cell markers to define cell types. In addition, the profiling data obtained still represents the averaged expression from a group of cells. Important features like inter-cell heterogeneity and cell subtypes may be masked in these population-averaged measurements.
It has been a longstanding goal in biology to be able to define all of the individual cells in a tissue of interest, the state each cell is in, and what cellular processes are active in each of these cells. The results of such an experiment should match the in vivo state as much as possible. Single cell RNA-sequencing (scRNA-seq)11, 12 comes close to this ideal through mRNA sequencing at single-cell resolution, representing a fundamentally new method for the comprehensive analysis of individual cells. It allows for the characterization of cell identity independent of predefined markers or assumptions regarding cell hierarchies. scRNA-seq generates data that allows the investigator to interrogate dynamic cellular process such as development, differentiation and disease pathogenesis, at a scale that has simply never been possible. In this review, we will discuss the development of scRNA-seq techniques and summarize potential applications in kidney disease investigation.
Development of scRNA-seq Protocols
All scRNA-seq techniques share several common steps: 1) single cell isolation, 2) mRNA capture, 3) cell lysis, 4) reverse transcription of mRNA, 5) amplification, 6) cDNA library generation and 7) next generation sequencing (Figure 1). The primary challenge is how to prepare complex cDNA libraries from the minute amount of mRNA in a single cell. A mammalian cell contains about 10 picograms of total RNA. Only 10–20% of this is reverse transcribed regardless of the scRNA-seq protocol and as a consequence all protocols utilize an RNA amplification step. Since the first scRNA-seq paper published in 2009 by Tang et al,13 numerous technical improvements have been made in both RNA amplification and library generation. Examples of such protocols using PCR-based amplification include STRT-seq,14 Smart-seq,15 Smart-seq2,16 SC3-seq,17 DropSeq,18 and SeqWell.19 An alternative approach to amplify mRNA utilizes in vitro transcription (IVT), and was developed and incorporated into CELL-Seq,20 CELL-Seq2,21 MARS-Seq22 and InDrops.23 The strengths and weaknesses of these methods have been recently reviewed.24
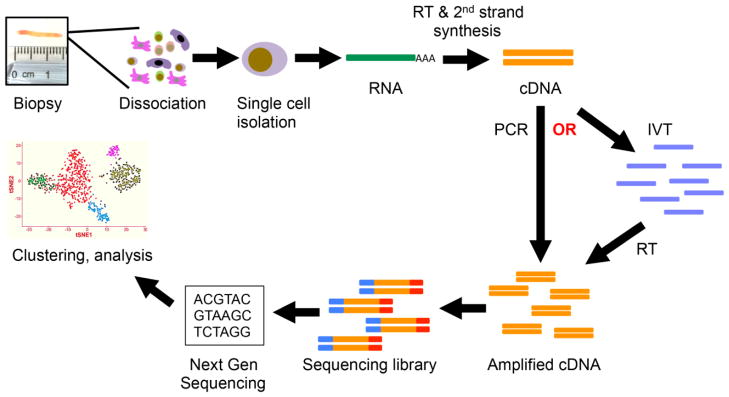
Figure 1. Applying scRNA-seq to human kidney biopsy analysis.
A kidney biopsy is first dissociated into a single cell suspension reflecting all cell types present in the biopsy. These cells are coencapsulated with lysis buffer and an oligo-dT containing primer. RNA is isolated through oligo-dT binding, and reverse transcribed (RT) with second strand synthesis into cDNA. This cDNA then can be amplified wither through PCR, or through in vitro transcription (IVT) followed by RT and second strand synthesis. Primers are then added to prepare the library for next generation sequencing. The sequencing data is analyzed to generate a digital count matrix reflecting the gene expression present in every cell from the biopsy. This can be analyzed in many different ways, one of them being cell clustering through dimensionality reduction. This groups cells by an unsupervised clustering approach which compares each cell’s transcriptome and clusters those that are similar. Cell types can then be classified by examining known marker gene expression in each cluster.
One complication of amplifying minute amounts of RNA is introduction of artifact. For example, some mRNA molecules may be amplified to a greater degree than others, leading to an artifactual overrepresentation of these molecules in the final library compared to their abundance in the original sample. Unique molecular identifiers (UMI)25 remove these amplification-associated artifacts and represent a critical advance in scRNA-Seq development. UMIs are random sequences of bases (or barcodes) that are used to label each transcript before library amplification. After sequencing, reads that have different barcodes represent different original molecules. This allows for the true quantification of unique mRNA molecules. During bioinformatics analysis, two reads that align to the same location and have the same UMI represent PCR duplicates. These artifactual PCR duplicates are tracked and collapsed during downstream analysis. The approach is summarized in Figure 2. This technology was first integrated in STRT-seq,25 and later inherited by CELL-Seq2,21 DropSeq,18 InDrops23 and SeqWell.19
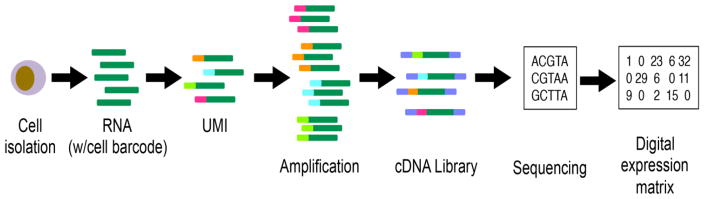
Figure 2. Unique Molecular Identifiers (UMI) to correct for amplification bias.
Creation of a single cell cDNA library requires amplication of pictograms of cDNA by many orders of magnitude. This is accomplished either by PCR or by in vitro transcription. Either way, bias is introduced if some transcripts undergo preferential amplification. To correct for this, a unique molecular identifier (UMI, which is essentially an oligonucleotide barcode, is added to each cDNA during reverse transcription and before amplification. Once the library is sequenced, these UMIs enable sequencing reads to be assigned to individual transcript molecules. If there is more than one read with the same UMI that maps to the same gene, this represents amplification bias and is collapsed into a single read informatically.
Recent technological advances in robotics, microfluidics and reverse emulsion droplets have dramatically increased assay throughput from hundreds of cells to tens of thousands of cells per experiment. These advances have actually scaled exponentially over the last 8 years - much faster than Moore’s law.26 The first single cell transcriptome was published in 2009. By 2011, 100’s of single cell transcriptomes were being published. By 2014, articles were describing thousands of single cell transcriptomes and today studies with sample sizes over 100,000 have been published.26 This dramatically increased scale has increased the power of the single-cell approach but also increased complexity of experimental design and data interpretation. We illustrate some of these issues in the next section by comparing plate-based and microfluidic-based scRNASeq assays.
Plate/tube-based scRNA-Seq
Constructing a cDNA library from a single cell is fundamentally similar to creating a cDNA library from a large group of cells. The main difference is that each cell yields an individual library rather than one library from many cells. The original method developed by Tang et al13, 27 used manual cell transfer of individual cells into individual PCR tubes making it difficult to scale up to hundreds of cells. Plate-based scRNA-seq such as STRT-seq significantly improved efficiency by transferring cells into a 96-well plate through a custom-built semi-automated cell picker.14 STRT-seq also introduced cell barcoding to cDNA libraries, which allowed the tracking of cell origin for each mRNA molecule throughout downstream analysis. Thus pooling and multiplexed analysis of all 96 samples at one time increased efficiency and economy. This important advance was achieved using a “template switching” technique. This strategy relies on the Moloney Murine Leukemia Virus (MMLV) reverse transcriptase. This transcriptase has intrinsic terminal transferase activity and adds several cytosines at the 3′ terminus of the cDNA first strand molecule. A template switching oligo (TS-oligo) with guanosine bases anneals to the cytosines, and the reverse transcriptase can switch templates and incorporate new sequences (in the TS-oligo) at both ends of the new cDNA molecule. By encoding different barcodes on the template switching oligonucleotide in each well of the 96-well plate, unique barcodes are added to each library (each cell). Libraries can then be pooled for sequencing and separated bioinformatically after sequencing. SMART-seq uses a similar template-switching strategy as STRT-seq but has enhanced ability to capture full-length transcripts.15 SMART-seq2 further improves cDNA yield and optimized reverse transcription, template switching and PCR amplification.16 Finally, Cell-Seq increased efficiency and accuracy by incorporating in vitro transcription amplification and multiplexing.20 CELL-Seq2 further enhanced the sensitivity and improved the quality of the data with the introduction of UMIs.21 All of these approaches require multiple steps and remain relatively laborious.
Plate and tube based scRNA-seq are also costly – about $5 – $10/cell for library creation. Modifications have been made to reduce the cost while preserving the capacity of scRNA-seq. Good examples include STRT-seq, SMART-seq and CELL-Seq adapted for use with the integrated fluidic circuits (IFC) in the Fluidigm C1 system.21, 25, 28 The Fluidigm C1 system can capture single cells in 96 nanoliter chambers, reducing cost. The autoprep system in Fluidigm C1 also allows user to simply load the cell suspension onto the chip without relying on FACS sorting, reducing labor. However, the cell capture chamber is a fixed size, which may bias against capture of cells with different sizes and frequently many chambers fail to capture a cell. Moreover, the cost of the Fluidigm C1 system remains high at about $3.5/cell. On the other hand, plate-based scRNA-seq provides unparalled transcript detection sensitivity. One recent study concluded that plate-based scRNA-seq can detect two-fold more genes per cell than microfluidic based scRNA-seq.29
Microfluidic based scRNA-Seq
New microfluidic droplet technologies were reported in 2015 that allow co-encapsulation of a cell, barcoded DNA oligonucleotides and cell lysis buffer within a tiny droplet of about 2 nanoliters.18, 23 This dramatically reduces the amount of RT reaction buffer used while increasing throughput. Droplets are one thousand times smaller than volumes in a traditional plate/tube based scRNA-seq assay and so these approaches are highly scalable allowing for parallel processing of thousands or tens of thousands of cells within an hour.
DropSeq
DropSeq, developed by Macosko et al, relies on two design concepts: a microfluidic chip and barcoded beads.18 The microfluidic chip allows co-flow of oil, cells, and barcoded beads to generate millions of nanoliter-size droplets within an hour. Cells and barcoded beads are randomly co-encapsulated in the droplets, resulting in thousands of droplets containing one cell and one bead.18 Each unique oligonucleotide sequence carried on DropSeq beads has four functions: 1) A primer handle for PCR amplification, 2) a cell barcode to tag each individual cell, 3) a UMI to tag each individual transcript in a cell, and 4) an oligo-dT to capture mRNA from each cell.18 Library construction in DropSeq consists of template-switching RT followed by PCR amplification. Due to the inexpensive and high throughput nature of DropSeq, it has become a widely used technique. There are publicly available resources, videos and discussion groups for DropSeq. To date, DropSeq has been successfully used to study the retina,18,30 and brain organoids to list just a few examples.31
Despite the widespread use of DropSeq there are limitations to this approach. It has a low cell capture efficiency, capturing only about 5% of input cells, making it unsuitable for analysis of precious clinical samples where the cell number is limited (e.g. human kidney biopsy). Another limitation is the low percentage of sequenced cDNA ‘reads’ that map to the reference genome. On average only 60%–80% of high quality raw reads will finally map to the genome, of which more than 30% are from cells that co-encapsulated with low quality beads (Malone, Wu and Humphreys, unpublished). The data from these beads must then be removed during downstream analysis thus further reducing the number of mapped reads available for final analysis. This increases sequencing costs due to the need to sequence at high depth. Finally, like other microfluidic techniques, DropSeq can only detect the top 20% most abundant transcripts and therefore has relatively low sensitivity to detect average or low expressed genes. Since many signaling molecules and transcription factors are expressed at low levels, DropSeq will not be able to detect many of them.
InDrops
InDrops is another microfluidic based scRNA-seq established by Klein et al23 and published at the same time as DropSeq. Its unique advantage is its capture rate. It can barcode 60%–90% of the total input cells. This is achieved by use of a deformable hydrogel bead containing the barcoded oligos, allowing very dense packing and synchronization of hydrogel release with droplet formation. InDrops is therefore a good choice for high throughput scRNA-seq analysis of samples containing a smaller number cells. Another feature of InDrops is that it adapts the IVT amplification method from CELL-Seq for library construction, minimizing amplification artifact.23 InDrops has been applied across different species and conditions. For example, Maayan et al has successfully used InDrops to compare the transcriptomes from mouse and human pancreas.32 Briggs et al applied InDrops to compare motor neurons generated by two differentiation protocols and revealed that the mature cell state can be reached via multiple differentiation paths.33 Limitations of the InDrops approach include higher technical skill required to pack the hydrogels in order to ensure regular release during the microfluidic droplet capture step, and library construction is somewhat more laborious.
A major advantage of both DropSeq and InDrops is the very low cost per cell. According to Macosko et al, reagent cost for DropSeq can be as low as 6 cents per cell, >100-fold lower than the plate-based scRNA-seq. The cost for InDrops is approximately 4 cents per cell. Cost savings are achieved because the RT reaction occurs in nanoliter droplets, reducing reagent volume requirements per cell. Costs are further reduced by the parallel processing of thousands of cells during cDNA library preparation allowed by the barcoding approach used by both DropSeq and InDrops.
10x Genomics and Illumina/BioRad
10X Genomics sells a microfluidic encapsulation machine called Chromium that can barcode 100 – 80,000 cells in 10 minutes with a 65% cell capture rate, at cost of only $0.15 – $1 per cell. The Chromium system has caught on rapidly among scientists because it is relatively easy to operate, can generate many thousands of single cell libraries in a short time frame and comes with a custom bioinformatics pipeline that simplifies analysis. This system is well suited to scRNA-seq of human kidney because of its high capture efficiency. This system is currently three to four-fold more expensive than DropSeq or InDrops, but the level of technical support provided is much greater – another benefit for labs new to scRNA-seq. Last year, Illumina announced their partnership with BioRad to bring a new high throughput scRNA platform. This system utilizes BioRad’s droplet technology (ddSEQ™ Single-Cell Isolator system) and Illumina’s Nextera library preparation system (SureCell WTA 3′ Library Prep Kit), and aims to separate and barcode 10,000 individual cells at $1 per cell, performed in a matter of hours. Both 10X Genomics and Illumina/BioRad also provide analytical software or computing space, a major asset for smaller labs that lack access to sufficient computing power.
Data analysis
Bioinformatic analysis of scRNA-seq data is computationally intensive. A detailed description of a typical scRNA-seq bioinformatic workflow is beyond the scope of this review but we will summarize the major steps. The initial steps are similar to bulk RNA-seq data analysis with the major difference being that UMIs and cell barcodes must be processed. First low-quality reads are removed and reads are mapped against the reference genome. Mapped reads are assigned to cell barcodes, and transcript counts for each gene are compiled for each cell. The resulting digital expression matrix represents an enormous dataset. Even a relatively simple DropSeq experiment might yield data for 5,000 cells, with 2,000 unique genes detected in each cell at specific expression levels.
Transforming the enormous amount of data produced by a single cell experiment into a result that is interpretable and informative is challenging and remains an area of rapid development. Dimensionality refers to the degree of variability between data points within a dataset. With thousands of genes in thousands of cells scRNA-seq datasets have very high dimensionality that must be simplified in order to detect the most significant variables in the dataset. Dimensionality reduction and unsupervised clustering methods process this high dimensional single cell data and can reveal cell type heterogeneity and diversity and other informative relationships between cells in the sample. Stochastic Neighbor Embedding (t-SNE) is a dimensionality reduction method that is suitable for visualizing high throughput scRNA-seq data. The Pe’er lab further developed a t-SNE machine learning algorithm and created viSNE which has been tested in their leukemia scRNA-seq dataset.34 More recently, ZIFA,35 a new dimensionality reduction approach, showed better performance on zero-inflated scRNA-seq data. In addition to dimensionality reduction, unsupervised graph-based clustering methods have also been developed to classify cell subpopulations and to identify markers responsible for the difference across subpopulations (e.g. SNN-Cliq36 and PhenoGraph37). Often investigators need to test multiple clustering algorithms on the same dataset in order to determine which one yields the most meaningful results for that particular experiment.
The development of machine learning tools has allowed for the ordering of cells along a trajectory corresponding to a biological process such as differentiation. This can be accomplished by a number of bioinformatics programs such as Monocle,38 Wishbone,39 Wanderlust,40 and DPT.41 Construction of cell trajectories with these tools can help to reveal markers for unrecognized intermediate cell states, genes regulating cell fate decisions, and cell subtypes. Network analysis on gene pairs derived from these programs further aids in elucidating gene regulatory dynamics during development, differentiation and disease progression. This kind of analysis will be extremely useful in nephrology research where the complex inter-cellular interactions involved in the normal function and pathophysiology of the kidney are yet to be fully elucidated.
Examples of scRNA-Seq in kidney research
To date, the majority of published scRNA-seq work is in the fields of neuroscience, stem cells and cancer. For example, Tirosh and colleagues performed scRNA-seq on 4,645 cells isolated from 19 patients with malignant melanoma.42 They discovered subsets of malignant cells - one defined by high expression of microphthalmia-associated transcription factor (MITF) while another expressed high levels of the receptor tyrosine kinase AXL. The AXL–high subgroup expressed other genes known to reflect resistance to targeted therapy – suggesting that these cells might represent dormant, therapy-resistant cells within the tumor. This AXL-high subgroup could not be detected through traditional bulk RNA-sequencing which emphasizes the power of scRNA-seq to reveal otherwise hidden cell types and states. Finally, the AXL-high subgroup was enriched in patients that had received targeted therapy – suggesting that indeed this was a dormant, therapy resistant group of cancer cells. This paper shows how scRNA-seq can help predict treatment outcomes and guide therapy.
scRNA-seq studies in nephrology are only just beginning to emerge. Brunskill et al profiled 235 cells from E11.5, E12.5 kidneys and P4 renal vesicles to unravel the early molecular mechanisms of renal precursors patterning.43 They provided important evidence that (at single cell resolution) initial kidney organogenesis involves a multi-lineage priming process. These results were produced using the Fluidigm C1 platform, and the SMART-seq library preparation strategy (full length RNA-seq) at high sequencing depth (2.6 million reads/cell) enabling investigation of transcript variation as well as non-coding RNA. Lu et al studied glomerular mesangial cells (n=33) using the Fluidigm C1 platform and found that the mesangial cell population is surprisingly heterogeneous.44 In this study, the authors also established a core set of markers for the study of mesangial cells.
Two groups have implemented scRNA-seq for the investigation of human kidney disease. Der et al isolated 361 renal biopsy cells from patients with lupus nephritis and profiled them on Fluidigm C1.45 They correlated clinical parameters (clinical scores of chronicity, proteinuria, and glomerular IgG deposition) and response to treatment at 12 months post biopsy with interferon responsive genes in tubular cells. A major limitation to this study was the lack of healthy kidney tissue as a control group. Kim et al used the Fluidigm C1 system46 to profile cells from paired samples of primary renal cell carcinoma (RCC) and metastatic RCC using scRNA-seq. They discovered cell heterogeneity between tumor sites (primary and metastatic) and within each site. This enabled them to develop a combinatorial therapeutic regimen that targeted two different cellular pathways active in either the primary or metastatic location. Based on scRNA-seq data, they proposed single-cell analysis-driven therapeutic strategy, which can be easily applied to other fields.
Major Limitations
The scRNA-seq studies described above represent important first results in nephrology research but they also highlight several limitations. First, the low throughput profiling using Fluidigm C1 system precludes the study of rare cell types. The Der et al experiments highlight this issue as two important cell types, the podocyte and mesangial cell, were not identified.45 Even though reagent cost has been reduced The Fluidigm C1 platform was a major advance in the field but cost per cell at $3.5 still prevents upscaling of experiments to thousands of cells.
Dissociation protocols have not been systematically examined between studies. Most dissociation protocols use various proteolytic enzymes incubated with tissue at body temperature. Enzymatic dissociation protocols usually compromise cell viability and so remain a concern with respect to the alteration of the transcriptional profile of the cells to be studied. We believe this to be one of the greatest limitations of scRNA-seq. Current methods lead to significant cell loss due to cell death and divergence from in vivo gene expression patterns. The presence of cell fragments and ambient RNA can further compromise microfluidic scRNA-seq by increasing noise substantially. To circumvent these limitations, many laboratories currently include a “clean-up” step after proteolytic dissociation. This consists of either a FACS purification, where only live cells are collected, or a density gradient centrifugation step which separates whole cells from fragments and ambient RNA. These approaches are summarized in figure 3. While simple wash steps may also achieve the same goal, in our experience these steps may compromise cell integrity without eliminating cell fragments (Malone, Wu and Humphreys, unpublished observations).
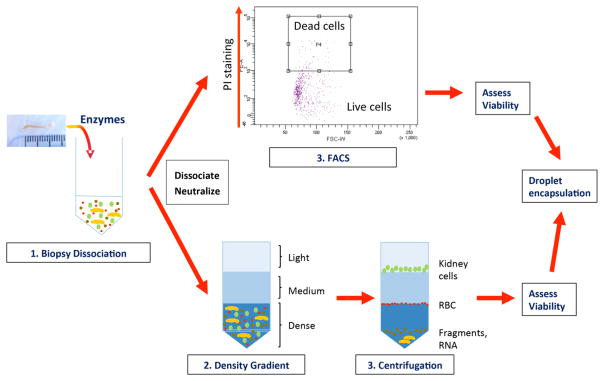
Figure 3. Dissociation and cleanup prior to droplet encapsulation.
Proteolytic dissociation of sold tissues such as kidney leads to the presence of many cell fragments and ambient RNA that must be removed prior to droplet encapsulation. This can be achieved by FACS purification, for example including propidium iodide (PI) which will bind to nucleic acids in cell fragments and dead cells. The PI-negative fraction is collected and subject to scRNA-seq. Alternatively, differential gradient separation can be used to separate whole cells from fragments because of their lower density. In either case, cell viability is assessed after the cleanup step (for example by Trypan blue exclusion) prior to droplet encapsulation.
Kidney Transplantation: Translational research potential of scRNA-sequencing
One of the major barriers to the application of scRNA-seq to human kidney research is the relative difficulty of obtaining live kidney tissue by way of biopsy or nephrectomy. An area of nephrology that could overcome this limitation is transplantation nephrology. The relatively easy access to kidney transplant tissue and high numbers of biopsies performed in the transplant clinic lends itself nicely to this technology. Gene expression analysis has been developed as a clinical diagnostic test for allograft rejection and fibrosis in kidney transplantation.47, 48 These methods use whole tissue samples to extract RNA and use microarray technology. Much of this work has been aimed at determining transcript sets that define different transplant pathologies, such as acute cellular rejection and antibody mediated rejection (AMR).49–52 By comparing the transcriptional profiles of hundreds of biopsies with different histologic diagnoses a classifier can be defined that can predict the tissue diagnosis.
Many interesting findings have come out of this work that have helped advance the field of transplant nephrology, in particular, the diagnosis and management of rejection. For example, AMR is now recognized as a major cause of late allograft loss.53 AMR is also difficult to diagnoses and histology is not sensitive. Microarray analysis of rejecting tissue has a role in helping the clinician make a diagnosis of rejection when histology is ambiguous. From a biological point of view his work has highlighted some of the pathways active during rejection episodes. In AMR endothelial cell transcript expression is increased, as are NK cell transcripts and IFNG-inducible genes. From a mechanistic point of view these data make sense as antibody binding to endothelial cells is the hallmark of AMR.
Despite an improved understanding of the mechanisms of rejection, these analyses only describe the ‘average’ of the genes expressed in the biopsy sample. A biopsy sample that contains a diverse set of cell types from immune cells to kidney cells. Bulk transcription analysis will miss or underestimate differentially expressed genes that may be important in disease but expressed at low level. These studies also lack the ability to characterize new cell types or cell types previously thought not to play a role in transplant rejection or pathology. Furthermore, the microarray bulk RNA approach lacks to resolution required to understand the subtle differences in disease pathology that must occur within the AMR setting. For example, it is known that AMR can occur with and without C4d deposition and that acute AMR does not always occur despite the presence of donor specific antibodies (DSA). Subtle molecular and cellular differences must exist to explain these clinical findings.
Single cell analysis of a rejecting kidney transplant has the potential to greatly increase our understanding of the mechanisms driving rejection, in particular the subtleties explaining the histologic variation seen in AMR and the hugely variable response to AMR treatment. It will likely lead to a better understanding of the immune cells involved and the pathways driving the alloimmune response. Furthermore, novel immune cell targets maybe revealed that might add to our current limited armamentarium of treatments for rejection.
On the Horizon: Future Developments
Current high throughput scRNA-seq technologies are likely to have a tremendous impact in nephrology. The ability to comprehensively define all cell types and states may fundamentally change the way we conceptualize heterogeneous diseases such as AKI, CKD and transplant rejection, providing better diagnostic tools, prognostic biomarkers and signaling pathways amenable to therapeutic targeting. Very recent droplet based scRNA-seq work in other tissues such as pancreas, retina and brain suggests that this analysis in kidney will almost certainly identify subpopulations, novel cell types and unappreciated heterogeneity.18, 23, 30–32 A gene expression atlas or systematic molecular classification of the kidney is an expressed goal of the NIDDK Kidney Precision Medicine Project54 and will inform our understanding of kidney function and guide future therapeutics in disease. These same approaches will also guide improvements in kidney organoid differentiation from pluripotent stem cells.
Sources of human kidney tissue are unpredictable and sample size is very small. Ideally these valuable tissue samples would be stored for later study without altering cell viability or expression patterns. However, there is an absence of protocols for the storage of tissue suitable for subsequent single cell analysis. The rapid emergence of nucleus RNA-seq may prove useful for the analysis of banked tissue samples where whole cell viability may be compromised. Two such methods include Div-Seq55 and DroNc-seq56 which appear to promise many of the advantages of current microfluidic approaches with the added ability to process stored tissue samples that are unfixed and frozen.
Conclusions
scRNA-seq is revolutionizing biology. There is enormous potential for application of scRNA-seq to kidney biopsies for an improved understanding of renal disease pathogenesis. We suggest that the field is developing so rapidly that scRNA-seq may become a routine component of human kidney biopsy analysis within a decade. Until then more kidney-focused laboratories should consider how they might apply scRNA-seq to better understand kidney development, normal physiology and disease.
Acknowledgments
Work in the Humphreys laboratory is supported by the NIH/NIDDK DK107374, DK103740 andDK104308 and by an Established Investigator Award of the American Heart Association.
Footnotes
Conflict of Interest: All authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Denic A, Mathew J, Lerman LO, Lieske JC, Larson JJ, Alexander MP, Poggio E, Glassock RJ, Rule AD. Single-Nephron Glomerular Filtration Rate in Healthy Adults. N Engl J Med. 2017;376:2349–2357. doi: 10.1056/NEJMoa1614329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Supavekin S, Zhang W, Kucherlapati R, Kaskel FJ, Moore LC, Devarajan P. Differential gene expression following early renal ischemia/reperfusion. Kidney Int. 2003;63:1714–1724. doi: 10.1046/j.1523-1755.2003.00928.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Xiong Y, Huang XR, Tang P, Yu X, Lan HY. Identification of Genes Associated with Smad3-dependent Renal Injury by RNA-seq-based Transcriptome Analysis. Sci Rep. 2015;5:17901. doi: 10.1038/srep17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakagawa S, Nishihara K, Miyata H, Shinke H, Tomita E, Kajiwara M, Matsubara T, Iehara N, Igarashi Y, Yamada H, Fukatsu A, Yanagita M, Matsubara K, Masuda S. Molecular Markers of Tubulointerstitial Fibrosis and Tubular Cell Damage in Patients with Chronic Kidney Disease. PLoS One. 2015;10:e0136994. doi: 10.1371/journal.pone.0136994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JW, Chou CL, Knepper MA. Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J Am Soc Nephrol. 2015;26:2669–2677. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, Zhang P project G. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 7.Boerries M, Grahammer F, Eiselein S, Buck M, Meyer C, Goedel M, Bechtel W, Zschiedrich S, Pfeifer D, Laloe D, Arrondel C, Goncalves S, Kruger M, Harvey SJ, Busch H, Dengjel J, Huber TB. Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int. 2013;83:1052–1064. doi: 10.1038/ki.2012.487. [DOI] [PubMed] [Google Scholar]
- 8.Grgic I, Krautzberger AM, Hofmeister A, Lalli M, DiRocco DP, Fleig SV, Liu J, Duffield JS, McMahon AP, Aronow B, Humphreys BD. Translational profiles of medullary myofibroblasts during kidney fibrosis. J Am Soc Nephrol. 2014;25:1979–1990. doi: 10.1681/ASN.2013101143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Krautzberger AM, Sui SH, Hofmann OM, Chen Y, Baetscher M, Grgic I, Kumar S, Humphreys BD, Hide WA, McMahon AP. Cell-specific translational profiling in acute kidney injury. J Clin Invest. 2014;124:1242–1254. doi: 10.1172/JCI72126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grgic I, Hofmeister AF, Genovese G, Bernhardy AJ, Sun H, Maarouf OH, Bijol V, Pollak MR, Humphreys BD. Discovery of new glomerular disease-relevant genes by translational profiling of podocytes in vivo. Kidney Int. 2014;86:1116–1129. doi: 10.1038/ki.2014.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 13.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 14.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura T, Yabuta Y, Okamoto I, Aramaki S, Yokobayashi S, Kurimoto K, Sekiguchi K, Nakagawa M, Yamamoto T, Saitou M. SC3-seq: a method for highly parallel and quantitative measurement of single-cell gene expression. Nucleic Acids Res. 2015;43:e60. doi: 10.1093/nar/gkv134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gierahn TM, Wadsworth MH, 2nd, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, Dor Y, Regev A, Yanai I. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 26.Svensson V, Vento-Tormo R, Teichman SA. Moore’s Law in Single Cell Transcriptomics. 2017 ArXiv e-prints. [Google Scholar]
- 27.Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, Quake SR. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Molecular cell. 2017;65:631–643. e634. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A, Sanes JR. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166:1308–1323. e1330. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, Vetere A, Ryu JH, Wagner BK, Shen-Orr SS, Klein AM, Melton DA, Yanai I. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst. 2016;3:346–360. e344. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briggs James Alexander, VCL, Lee Seungkyu, Woolf Clifford J, Klein Allon, Kirschner Marc W. Mouse embryonic stem cells can differentiate via multiple paths to the same state. bioRxiv. 2017 doi: 10.7554/eLife.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierson E, Yau C. ZIFA: Dimensionality reduction for zero-inflated single-cell gene expression analysis. Genome Biol. 2015;16:241. doi: 10.1186/s13059-015-0805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu C, Su Z. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics. 2015;31:1974–1980. doi: 10.1093/bioinformatics/btv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, Finck R, Gedman AL, Radtke I, Downing JR, Pe’er D, Nolan GP. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Bendall S, Friedman N, Pe’er D. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol. 2016;34:637–645. doi: 10.1038/nbt.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bendall SC, Davis KL, Amir el AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 42.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin JR, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CG, Kazer SW, Gaillard A, Kolb KE, Villani AC, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jane-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A, Garraway LA. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunskill EW, Park JS, Chung E, Chen F, Magella B, Potter SS. Single cell dissection of early kidney development: multilineage priming. Development. 2014;141:3093–3101. doi: 10.1242/dev.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Y, Ye Y, Yang Q, Shi S. Single-cell RNA-sequence analysis of mouse glomerular mesangial cells uncovers mesangial cell essential genes. Kidney Int. 2017 doi: 10.1016/j.kint.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 45.Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, Kustagi M, Czuppa M, Izmirly P, Belmont HM, Wang T, Jordan N, Bornkamp N, Nwaukoni J, Martinez J, Goilav B, Buyon JP, Tuschl T, Putterman C. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight. 2017;2 doi: 10.1172/jci.insight.93009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim KT, Lee HW, Lee HO, Song HJ, Jeong da E, Shin S, Kim H, Shin Y, Nam DH, Jeong BC, Kirsch DG, Joo KM, Park WY. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. 2016;17:80. doi: 10.1186/s13059-016-0945-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Connell PJ, Zhang W, Menon MC, Yi Z, Schroppel B, Gallon L, Luan Y, Rosales IA, Ge Y, Losic B, Xi C, Woytovich C, Keung KL, Wei C, Greene I, Overbey J, Bagiella E, Najafian N, Samaniego M, Djamali A, Alexander SI, Nankivell BJ, Chapman JR, Smith RN, Colvin R, Murphy B. Biopsy transcriptome expression profiling to identify kidney transplants at risk of chronic injury: a multicentre, prospective study. Lancet. 2016;388:983–993. doi: 10.1016/S0140-6736(16)30826-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halloran PF, Reeve J, Akalin E, Aubert O, Bohmig GA, Brennan D, Bromberg J, Einecke G, Eskandary F, Gosset C, Duong Van Huyen JP, Gupta G, Lefaucheur C, Malone A, Mannon RB, Seron D, Sellares J, Weir M, Loupy A. Real time central assessment of kidney transplant indication biopsies by microarrays: The INTERCOMEX Study. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2017 doi: 10.1111/ajt.14329. [DOI] [PubMed] [Google Scholar]
- 49.Reeve J, Sellares J, Mengel M, Sis B, Skene A, Hidalgo L, de Freitas DG, Famulski KS, Halloran PF. Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2013;13:645–655. doi: 10.1111/ajt.12079. [DOI] [PubMed] [Google Scholar]
- 50.Halloran PF, Pereira AB, Chang J, Matas A, Picton M, De Freitas D, Bromberg J, Seron D, Sellares J, Einecke G, Reeve J. Potential impact of microarray diagnosis of T cell-mediated rejection in kidney transplants: The INTERCOM study. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2013;13:2352–2363. doi: 10.1111/ajt.12387. [DOI] [PubMed] [Google Scholar]
- 51.Sellares J, Reeve J, Loupy A, Mengel M, Sis B, Skene A, de Freitas DG, Kreepala C, Hidalgo LG, Famulski KS, Halloran PF. Molecular diagnosis of antibody-mediated rejection in human kidney transplants. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2013;13:971–983. doi: 10.1111/ajt.12150. [DOI] [PubMed] [Google Scholar]
- 52.Halloran PF, Pereira AB, Chang J, Matas A, Picton M, De Freitas D, Bromberg J, Seron D, Sellares J, Einecke G, Reeve J. Microarray diagnosis of antibody-mediated rejection in kidney transplant biopsies: an international prospective study (INTERCOM) American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2013;13:2865–2874. doi: 10.1111/ajt.12465. [DOI] [PubMed] [Google Scholar]
- 53.Matas A, Fieberg A, Leduc R, Cosio F, Gaston R, Mannon R, Cecka M, Rush D, Kasiske B, Gourishankar S, Connett JJG. Long-Term Death-Censored Graft Loss in the DeKAF Study. [abstract] Am J Transplant. 2016;16(suppl 3):16. [Google Scholar]
- 54.The NIDDK Kidney Precision Medicine Project (KPMP) [Accessed 05/29/2017]; https://wwwniddknihgov/research-funding/research-programs/kidney-precision-medicine-project-kpmp.
- 55.Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, Regev A. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Habib Naomi, AB, Avraham-Davidi Inbal, Burks Tyler, Choudhury Sourav R, Aguet Francois, Gelfand Ellen, Ardlie Kristin, Weitz David A, Rozenblatt-Rosen Orit, Zhang Feng, Regev Aviv. DroNc-Seq: Deciphering cell types in human archived brain tissues by massively-parallel single nucleus RNA-seq. bioRxiv. 2017 doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]